Gene
KWMTBOMO15047 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA004859
Annotation
PREDICTED:_N-acetylneuraminate_lyase-like_isoform_X1_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.366
Sequence
CDS
ATGATTGACTTTGAAACACGTGGTTTGATGCCCCCGGTCTTCACGCCGCTGAACGATGATCTCACAATCAACTATGCGGCTATACCTGGATATGCCAAATACATGGCGGAAGCTGGCTTCAAATGTGTACTCGTTGGTGGTACGACAGGCGAGCATATGTCTCTAAACGTAGCGGACAGGAAGAAAGTTATCGAAGAATGGGTGAAAGCGGGAAGGAGCACTGGTCTTCACATACAGGTACAAGTCGGTGGTGCACCACTCCCCGACGTGCAGGAATTGGCGGCGTTCTGTCAAAAAAATGGCGTTGGTTCGATTCTAACGCTACCGGAACTATACTTCAAACCGAGCAATGTCGATGAACTGGTGTCTTACGTTGCCCTGGTAGCCGGCGCGGCACCGAAATTGCCTGTGCTATATTACCACATTCCCAGCATGACTCGGGTTGAGATTAACATGCCTGCCTTCGTCACGGAGGCCTCAAAGAAAATACCCAATTTCGCAGGCATCAAGTTCACTTCGAACGATTTGAGCGAAGCGTCTCAAGTGCTTCGTGCCATGTCCGAGAAGAAGGCACTATTCCTTGGAGCGGACACGCTTTTAGCACCGGCGGCCCTGCTCGGCATAAAATCGAGCATCGGGACGTCTTTCAATCTCTTCCCGAAAATAGCTCACGCTATCCTGAGCGCTGTTCAGAATAAGGACGTGGAGACGGCGAGAGCCCTCCAGCACAAGCTGTGTCTGGCGATCGAATCATTGACCAAGGAAGGTCCGTGGGTGCCCGTTATGAAGGCCGGAATGGAAATCGTGACCGGAATCAGAGTTGGGCCGCCTTCGCTGCCTCAAAAGCCGCTTAGTTCGGAAGCCATAGAGCGGATTCGTGGCAGGCTCAGGGTGCTGGATGTAGCCTAG
Protein
MIDFETRGLMPPVFTPLNDDLTINYAAIPGYAKYMAEAGFKCVLVGGTTGEHMSLNVADRKKVIEEWVKAGRSTGLHIQVQVGGAPLPDVQELAAFCQKNGVGSILTLPELYFKPSNVDELVSYVALVAGAAPKLPVLYYHIPSMTRVEINMPAFVTEASKKIPNFAGIKFTSNDLSEASQVLRAMSEKKALFLGADTLLAPAALLGIKSSIGTSFNLFPKIAHAILSAVQNKDVETARALQHKLCLAIESLTKEGPWVPVMKAGMEIVTGIRVGPPSLPQKPLSSEAIERIRGRLRVLDVA
Summary
Similarity
Belongs to the DapA family.
Uniprot
H9J5R9
A0A2A4JFY8
I4DPJ4
A0A2A4JFM5
A0A194PFM7
A0A0N0PCS5
+ More
A0A3S2NC89 A0A212ETD9 A0A0L7L8A9 J3JYH0 F4X1Y9 D6WME9 E2BFR7 A0A195FJA2 A0A158P438 A0A151IUE1 A0A2A4JEY1 A0A151WF36 E9IK31 N6UI15 A0A0C9RWH4 K7J245 A0A2J7QQ19 A0A023F7H9 A0A067RJQ4 A0A2J7QPZ4 R4FL94 A0A0A9Z1Y6 A0A0K8SKB8 A0A1Y1KGM9 A0A151JBW8 A0A1Y1KH54 A0A232FEM4 A0A151WF58 K7IMV4 A0A158P437 A0A087ZVV2 A0A1B6DZ58 E2AJ86 F4X1Z0 A0A2H1W3A5 A0A1Y1KKF9 A0A195FJP7 A0A224XHH7 A0A1B6LID4 A0A1B6IYA4 A0A0C9QMI7 A0A1L8DCV7 A0A0N0BGL6 H9J5M0 A0A1Y1KN37 A0A026WBS3 A0A2A3EF84 A0A3S2L1T7 A0A336M102 A0A1B6I5W9 E0VUC4 A0A2J7QQ05 A0A0T6ATU1 A0A1Y1KBI2 A0A1W4X969 A0A1W4X7X0 A0A0P4VHC9 Q16WA2 A0A1S4FMA1 A0A182GZ29 A0A195CNB7 A0A182QP80 R4WNH6 A0A154PSC7 A0A182T0U9 A0A1B0CHZ1 A0A310SPK1 A0A182NHC9 A0A084VF66 A0A2W1BRW0 A0A1L8DBH0 A0A182M3E0 A0A1Q3FDN6 A0A182JMC6 B0VZR3 A0A232EGK8 A0A336M1U9 A0A182WIY3 A0A1B0D3L4 A0A1B6CQ74 A0A182PA02 T1HX93 A0A182FG30 A0A0L7RKK5 A0A182X6K7 A0A182KS08 T1DNX9 Q7PTG5 A0A1Y1LHN7 W5JKQ9
A0A3S2NC89 A0A212ETD9 A0A0L7L8A9 J3JYH0 F4X1Y9 D6WME9 E2BFR7 A0A195FJA2 A0A158P438 A0A151IUE1 A0A2A4JEY1 A0A151WF36 E9IK31 N6UI15 A0A0C9RWH4 K7J245 A0A2J7QQ19 A0A023F7H9 A0A067RJQ4 A0A2J7QPZ4 R4FL94 A0A0A9Z1Y6 A0A0K8SKB8 A0A1Y1KGM9 A0A151JBW8 A0A1Y1KH54 A0A232FEM4 A0A151WF58 K7IMV4 A0A158P437 A0A087ZVV2 A0A1B6DZ58 E2AJ86 F4X1Z0 A0A2H1W3A5 A0A1Y1KKF9 A0A195FJP7 A0A224XHH7 A0A1B6LID4 A0A1B6IYA4 A0A0C9QMI7 A0A1L8DCV7 A0A0N0BGL6 H9J5M0 A0A1Y1KN37 A0A026WBS3 A0A2A3EF84 A0A3S2L1T7 A0A336M102 A0A1B6I5W9 E0VUC4 A0A2J7QQ05 A0A0T6ATU1 A0A1Y1KBI2 A0A1W4X969 A0A1W4X7X0 A0A0P4VHC9 Q16WA2 A0A1S4FMA1 A0A182GZ29 A0A195CNB7 A0A182QP80 R4WNH6 A0A154PSC7 A0A182T0U9 A0A1B0CHZ1 A0A310SPK1 A0A182NHC9 A0A084VF66 A0A2W1BRW0 A0A1L8DBH0 A0A182M3E0 A0A1Q3FDN6 A0A182JMC6 B0VZR3 A0A232EGK8 A0A336M1U9 A0A182WIY3 A0A1B0D3L4 A0A1B6CQ74 A0A182PA02 T1HX93 A0A182FG30 A0A0L7RKK5 A0A182X6K7 A0A182KS08 T1DNX9 Q7PTG5 A0A1Y1LHN7 W5JKQ9
Pubmed
19121390
22651552
26354079
22118469
26227816
22516182
+ More
21719571 18362917 19820115 20798317 21347285 21282665 23537049 20075255 25474469 24845553 25401762 26823975 28004739 28648823 24508170 20566863 27129103 17510324 26483478 23691247 24438588 28756777 20966253 12364791 14747013 17210077 20920257 23761445
21719571 18362917 19820115 20798317 21347285 21282665 23537049 20075255 25474469 24845553 25401762 26823975 28004739 28648823 24508170 20566863 27129103 17510324 26483478 23691247 24438588 28756777 20966253 12364791 14747013 17210077 20920257 23761445
EMBL
BABH01025065
BABH01025066
NWSH01001636
PCG70618.1
AK403635
BAM19834.1
+ More
PCG70619.1 KQ459604 KPI92181.1 KQ460594 KPJ13942.1 RSAL01000235 RVE43727.1 AGBW02012595 OWR44747.1 JTDY01002387 KOB71521.1 BT128298 AEE63256.1 GL888558 EGI59525.1 KQ971343 EFA04261.2 GL448039 EFN85455.1 KQ981523 KYN40483.1 ADTU01008424 KQ980964 KYN11012.1 PCG70617.1 KQ983227 KYQ46426.1 GL763924 EFZ18997.1 APGK01035366 APGK01035367 KB740923 ENN78297.1 GBYB01013345 JAG83112.1 NEVH01012087 PNF30663.1 GBBI01001520 JAC17192.1 KK852639 KDR19640.1 PNF30658.1 GAHY01001523 JAA75987.1 GBHO01005141 GBHO01005140 GDHC01021560 JAG38463.1 JAG38464.1 JAP97068.1 GBRD01012074 GBRD01012073 GDHC01015486 JAG53751.1 JAQ03143.1 GEZM01087347 JAV58745.1 KQ979110 KYN22602.1 GEZM01087346 JAV58746.1 NNAY01000352 OXU28970.1 KYQ46425.1 GEDC01006332 GEDC01003561 JAS30966.1 JAS33737.1 GL439967 EFN66526.1 EGI59526.1 ODYU01006029 SOQ47513.1 GEZM01081089 JAV61892.1 KYN40482.1 GFTR01004480 JAW11946.1 GEBQ01016525 JAT23452.1 GECU01015785 JAS91921.1 GBYB01004719 JAG74486.1 GFDF01009887 JAV04197.1 KQ435775 KOX74992.1 GEZM01081090 JAV61891.1 KK107293 EZA53418.1 KZ288262 PBC30397.1 RVE43726.1 UFQT01000393 SSX23925.1 GECU01025380 JAS82326.1 DS235785 EEB16980.1 PNF30659.1 LJIG01022832 KRT78489.1 GEZM01087348 JAV58744.1 GDKW01002751 JAI53844.1 CH477572 EAT38848.1 JXUM01098550 KQ564429 KXJ72292.1 KQ977513 KYN02246.1 AXCN02000896 AK417186 BAN20401.1 KQ435100 KZC14657.1 AJWK01012915 AJWK01012916 KQ760538 OAD59997.1 ATLV01012362 KE524785 KFB36610.1 KZ149918 PZC77822.1 GFDF01010410 JAV03674.1 AXCM01001459 GFDL01009354 JAV25691.1 DS231815 EDS35155.1 NNAY01004742 OXU17487.1 UFQT01000285 SSX22803.1 AJVK01002966 AJVK01002967 GEDC01028887 GEDC01021641 JAS08411.1 JAS15657.1 ACPB03006242 KQ414570 KOC71349.1 GAMD01002465 JAA99125.1 AAAB01008807 EAA03961.4 GEZM01055082 JAV73162.1 ADMH02001258 ETN63359.1
PCG70619.1 KQ459604 KPI92181.1 KQ460594 KPJ13942.1 RSAL01000235 RVE43727.1 AGBW02012595 OWR44747.1 JTDY01002387 KOB71521.1 BT128298 AEE63256.1 GL888558 EGI59525.1 KQ971343 EFA04261.2 GL448039 EFN85455.1 KQ981523 KYN40483.1 ADTU01008424 KQ980964 KYN11012.1 PCG70617.1 KQ983227 KYQ46426.1 GL763924 EFZ18997.1 APGK01035366 APGK01035367 KB740923 ENN78297.1 GBYB01013345 JAG83112.1 NEVH01012087 PNF30663.1 GBBI01001520 JAC17192.1 KK852639 KDR19640.1 PNF30658.1 GAHY01001523 JAA75987.1 GBHO01005141 GBHO01005140 GDHC01021560 JAG38463.1 JAG38464.1 JAP97068.1 GBRD01012074 GBRD01012073 GDHC01015486 JAG53751.1 JAQ03143.1 GEZM01087347 JAV58745.1 KQ979110 KYN22602.1 GEZM01087346 JAV58746.1 NNAY01000352 OXU28970.1 KYQ46425.1 GEDC01006332 GEDC01003561 JAS30966.1 JAS33737.1 GL439967 EFN66526.1 EGI59526.1 ODYU01006029 SOQ47513.1 GEZM01081089 JAV61892.1 KYN40482.1 GFTR01004480 JAW11946.1 GEBQ01016525 JAT23452.1 GECU01015785 JAS91921.1 GBYB01004719 JAG74486.1 GFDF01009887 JAV04197.1 KQ435775 KOX74992.1 GEZM01081090 JAV61891.1 KK107293 EZA53418.1 KZ288262 PBC30397.1 RVE43726.1 UFQT01000393 SSX23925.1 GECU01025380 JAS82326.1 DS235785 EEB16980.1 PNF30659.1 LJIG01022832 KRT78489.1 GEZM01087348 JAV58744.1 GDKW01002751 JAI53844.1 CH477572 EAT38848.1 JXUM01098550 KQ564429 KXJ72292.1 KQ977513 KYN02246.1 AXCN02000896 AK417186 BAN20401.1 KQ435100 KZC14657.1 AJWK01012915 AJWK01012916 KQ760538 OAD59997.1 ATLV01012362 KE524785 KFB36610.1 KZ149918 PZC77822.1 GFDF01010410 JAV03674.1 AXCM01001459 GFDL01009354 JAV25691.1 DS231815 EDS35155.1 NNAY01004742 OXU17487.1 UFQT01000285 SSX22803.1 AJVK01002966 AJVK01002967 GEDC01028887 GEDC01021641 JAS08411.1 JAS15657.1 ACPB03006242 KQ414570 KOC71349.1 GAMD01002465 JAA99125.1 AAAB01008807 EAA03961.4 GEZM01055082 JAV73162.1 ADMH02001258 ETN63359.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000283053
UP000007151
+ More
UP000037510 UP000007755 UP000007266 UP000008237 UP000078541 UP000005205 UP000078492 UP000075809 UP000019118 UP000002358 UP000235965 UP000027135 UP000215335 UP000005203 UP000000311 UP000053105 UP000053097 UP000242457 UP000009046 UP000192223 UP000008820 UP000069940 UP000249989 UP000078542 UP000075886 UP000076502 UP000075901 UP000092461 UP000075884 UP000030765 UP000075883 UP000075880 UP000002320 UP000075920 UP000092462 UP000075885 UP000015103 UP000069272 UP000053825 UP000076407 UP000075882 UP000007062 UP000000673
UP000037510 UP000007755 UP000007266 UP000008237 UP000078541 UP000005205 UP000078492 UP000075809 UP000019118 UP000002358 UP000235965 UP000027135 UP000215335 UP000005203 UP000000311 UP000053105 UP000053097 UP000242457 UP000009046 UP000192223 UP000008820 UP000069940 UP000249989 UP000078542 UP000075886 UP000076502 UP000075901 UP000092461 UP000075884 UP000030765 UP000075883 UP000075880 UP000002320 UP000075920 UP000092462 UP000075885 UP000015103 UP000069272 UP000053825 UP000076407 UP000075882 UP000007062 UP000000673
PRIDE
Interpro
Gene 3D
ProteinModelPortal
H9J5R9
A0A2A4JFY8
I4DPJ4
A0A2A4JFM5
A0A194PFM7
A0A0N0PCS5
+ More
A0A3S2NC89 A0A212ETD9 A0A0L7L8A9 J3JYH0 F4X1Y9 D6WME9 E2BFR7 A0A195FJA2 A0A158P438 A0A151IUE1 A0A2A4JEY1 A0A151WF36 E9IK31 N6UI15 A0A0C9RWH4 K7J245 A0A2J7QQ19 A0A023F7H9 A0A067RJQ4 A0A2J7QPZ4 R4FL94 A0A0A9Z1Y6 A0A0K8SKB8 A0A1Y1KGM9 A0A151JBW8 A0A1Y1KH54 A0A232FEM4 A0A151WF58 K7IMV4 A0A158P437 A0A087ZVV2 A0A1B6DZ58 E2AJ86 F4X1Z0 A0A2H1W3A5 A0A1Y1KKF9 A0A195FJP7 A0A224XHH7 A0A1B6LID4 A0A1B6IYA4 A0A0C9QMI7 A0A1L8DCV7 A0A0N0BGL6 H9J5M0 A0A1Y1KN37 A0A026WBS3 A0A2A3EF84 A0A3S2L1T7 A0A336M102 A0A1B6I5W9 E0VUC4 A0A2J7QQ05 A0A0T6ATU1 A0A1Y1KBI2 A0A1W4X969 A0A1W4X7X0 A0A0P4VHC9 Q16WA2 A0A1S4FMA1 A0A182GZ29 A0A195CNB7 A0A182QP80 R4WNH6 A0A154PSC7 A0A182T0U9 A0A1B0CHZ1 A0A310SPK1 A0A182NHC9 A0A084VF66 A0A2W1BRW0 A0A1L8DBH0 A0A182M3E0 A0A1Q3FDN6 A0A182JMC6 B0VZR3 A0A232EGK8 A0A336M1U9 A0A182WIY3 A0A1B0D3L4 A0A1B6CQ74 A0A182PA02 T1HX93 A0A182FG30 A0A0L7RKK5 A0A182X6K7 A0A182KS08 T1DNX9 Q7PTG5 A0A1Y1LHN7 W5JKQ9
A0A3S2NC89 A0A212ETD9 A0A0L7L8A9 J3JYH0 F4X1Y9 D6WME9 E2BFR7 A0A195FJA2 A0A158P438 A0A151IUE1 A0A2A4JEY1 A0A151WF36 E9IK31 N6UI15 A0A0C9RWH4 K7J245 A0A2J7QQ19 A0A023F7H9 A0A067RJQ4 A0A2J7QPZ4 R4FL94 A0A0A9Z1Y6 A0A0K8SKB8 A0A1Y1KGM9 A0A151JBW8 A0A1Y1KH54 A0A232FEM4 A0A151WF58 K7IMV4 A0A158P437 A0A087ZVV2 A0A1B6DZ58 E2AJ86 F4X1Z0 A0A2H1W3A5 A0A1Y1KKF9 A0A195FJP7 A0A224XHH7 A0A1B6LID4 A0A1B6IYA4 A0A0C9QMI7 A0A1L8DCV7 A0A0N0BGL6 H9J5M0 A0A1Y1KN37 A0A026WBS3 A0A2A3EF84 A0A3S2L1T7 A0A336M102 A0A1B6I5W9 E0VUC4 A0A2J7QQ05 A0A0T6ATU1 A0A1Y1KBI2 A0A1W4X969 A0A1W4X7X0 A0A0P4VHC9 Q16WA2 A0A1S4FMA1 A0A182GZ29 A0A195CNB7 A0A182QP80 R4WNH6 A0A154PSC7 A0A182T0U9 A0A1B0CHZ1 A0A310SPK1 A0A182NHC9 A0A084VF66 A0A2W1BRW0 A0A1L8DBH0 A0A182M3E0 A0A1Q3FDN6 A0A182JMC6 B0VZR3 A0A232EGK8 A0A336M1U9 A0A182WIY3 A0A1B0D3L4 A0A1B6CQ74 A0A182PA02 T1HX93 A0A182FG30 A0A0L7RKK5 A0A182X6K7 A0A182KS08 T1DNX9 Q7PTG5 A0A1Y1LHN7 W5JKQ9
PDB
6ARH
E-value=4.35259e-44,
Score=447
Ontologies
KEGG
PATHWAY
GO
PANTHER
Topology
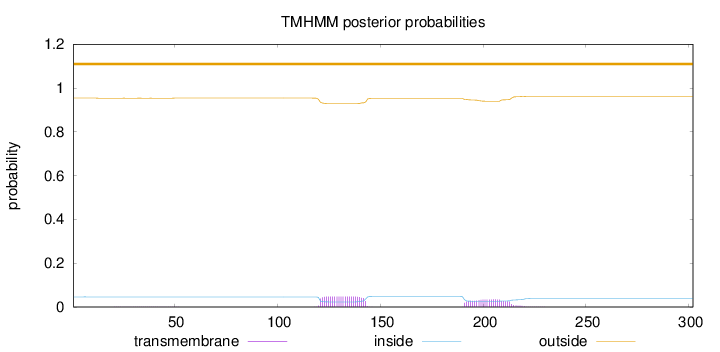
Length:
302
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.81136
Exp number, first 60 AAs:
0.01658
Total prob of N-in:
0.04602
outside
1 - 302
Population Genetic Test Statistics
Pi
253.523726
Theta
17.103109
Tajima's D
0.738558
CLR
0.630679
CSRT
0.593920303984801
Interpretation
Uncertain
