Gene
KWMTBOMO15032 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA004866
Annotation
PREDICTED:_insecticyanin-A-like_isoform_X1_[Amyelois_transitella]
Full name
Insecticyanin-A
+ More
Insecticyanin-B
Apolipoprotein D
Insecticyanin-B
Apolipoprotein D
Alternative Name
Blue biliprotein
Location in the cell
Extracellular Reliability : 1.644 Nuclear Reliability : 1.299
Sequence
CDS
ATGTTAATAAAACATTTAATATTTGCTGCCAGTTTTTGTGTTGTTCACGCATACTTCACTTTACCAGGAAAATGTCCGGCAGATGTTCAATTACAACAGGAATTCAGATTGGCTGATTTCTTCGGTAAATGGTATCAAGCTTATCACTATTCCAGCGATGAACAGCAGCAGAATAATTGCTCGGTCCTAGAGCTTCAGACCAAGCCCAGCGGCATATACCTGAACCAGTCAAGGATAGATAGGGGCTTGTTTCACCGTTATAGTATCGCTAAATTGGAGATACCGTCGAATTTAGAAGACGCTGCTAAATTAAATGTCAAGTTTTTCTTCGAAAACGCTCCTAGGAGACTGAGGATAAGAAGATATCCATTTTCCGTTTTGGCCACTAATTACCAATATTACGCTACCGTCTACACGTGTCAGTACAGTCCACTAACAGATAAGCATTTTATCTACATCTGGATTTTATCAAGGAATCCAATCTTGAACGATGTGTCGAAAGAATTAGCTACGAAGCCACTAAGTCAACTTGGCATTGACGTAACGAAGATCAAGAAAGACGATCTTACTAGATGCACTCCGAAATATTATGAAGATTCCCAAACTGAACCAATGACCTTTAGATATCCTGTACCAGTATAG
Protein
MLIKHLIFAASFCVVHAYFTLPGKCPADVQLQQEFRLADFFGKWYQAYHYSSDEQQQNNCSVLELQTKPSGIYLNQSRIDRGLFHRYSIAKLEIPSNLEDAAKLNVKFFFENAPRRLRIRRYPFSVLATNYQYYATVYTCQYSPLTDKHFIYIWILSRNPILNDVSKELATKPLSQLGIDVTKIKKDDLTRCTPKYYEDSQTEPMTFRYPVPV
Summary
Description
APOD occurs in the macromolecular complex with lecithin-transport and binding of bilin. Appears to be able to transport a variety of ligands in a number of different contexts (By similarity).
Subunit
Homotetramer.
Homodimer.
Homodimer.
Similarity
Belongs to the calycin superfamily. Lipocalin family.
Keywords
3D-structure
Bile pigment
Chromophore
Direct protein sequencing
Disulfide bond
Pigment
Secreted
Signal
Complete proteome
Glycoprotein
Lipid-binding
Pyrrolidone carboxylic acid
Reference proteome
Transport
Feature
chain Insecticyanin-A
Uniprot
H9J5S6
A0A2H1VQH8
A0A2W1BRV6
S4PVS5
A0A2A4JU62
A0A212EIU3
+ More
A0A194RM77 A0A2A4JTF5 A0A194PHH4 Q4SK77 H2MUK0 H2MUJ9 A0A3P9K5L3 A0A3P9K5M3 A0A3P9IV14 A0A3P9H6J3 A0A194PM29 A0A084WTA1 M4ATP2 A0A2U9BID1 A0A3B3DF18 A0A147AQ88 A0A3B4UH23 A0A087XJ49 A0A3B3U536 A0A3Q2P803 A0A2H1VSA5 A0A0L7KXZ8 A0A2W1BYB5 A0A3P9PEY2 A0A3P8W739 A0A3B3X9V5 A0A3Q1HHN3 Q00629 A0A194RHP6 A0A3Q0RF77 A0A3Q0RKG1 A0A2A4JTE9 I3KQQ7 A0A3Q1HDT0 A0A1E1WGA5 A0A3Q2VUM2 A0A3P9DDR8 P00305 Q24996 A0A3P8PHW7 A0A3Q4HDD4 G3PXV7 A0A3Q4BXK7 A0A159YQU9 A0A3Q3XJW8 H2SCU4 A0A0S7MDZ3 Q00630 A0A3Q1JNI9 A0A3B4FLM3 A3KEW8 A0A3B5A5P2 A0A3Q2D0T8 L5L1J2 W8C9C1 A0A315W1Y7 A0A3Q1KCJ4 G3PXW5 F1SQX9 A0A3Q1CF05 H2SBW8 A0A315W2H7 A0A1A8VAG0 A0A3Q2D080 A0A182J1F2 A0A3B4UFP1 A0A1A6HRT1 Q32KY0 A0A3Q0QX35 A0A182LEN1 F1MS32 A0A3P8U4T7 F5HKU4 A0A0K8UND9 L8IIL2 A0A182XUT6 A0A2C9GRE4 A0A1S3FFP1 D2HH45 G1LS67 C3YC60 F6XM13 P51910 A0A3Q0CSM2 A0A340XXN3 G3T8L4 A0A1D2MJS8 P23593 A0A3B5MND9 M0R4S2 A0A1D2MVQ6
A0A194RM77 A0A2A4JTF5 A0A194PHH4 Q4SK77 H2MUK0 H2MUJ9 A0A3P9K5L3 A0A3P9K5M3 A0A3P9IV14 A0A3P9H6J3 A0A194PM29 A0A084WTA1 M4ATP2 A0A2U9BID1 A0A3B3DF18 A0A147AQ88 A0A3B4UH23 A0A087XJ49 A0A3B3U536 A0A3Q2P803 A0A2H1VSA5 A0A0L7KXZ8 A0A2W1BYB5 A0A3P9PEY2 A0A3P8W739 A0A3B3X9V5 A0A3Q1HHN3 Q00629 A0A194RHP6 A0A3Q0RF77 A0A3Q0RKG1 A0A2A4JTE9 I3KQQ7 A0A3Q1HDT0 A0A1E1WGA5 A0A3Q2VUM2 A0A3P9DDR8 P00305 Q24996 A0A3P8PHW7 A0A3Q4HDD4 G3PXV7 A0A3Q4BXK7 A0A159YQU9 A0A3Q3XJW8 H2SCU4 A0A0S7MDZ3 Q00630 A0A3Q1JNI9 A0A3B4FLM3 A3KEW8 A0A3B5A5P2 A0A3Q2D0T8 L5L1J2 W8C9C1 A0A315W1Y7 A0A3Q1KCJ4 G3PXW5 F1SQX9 A0A3Q1CF05 H2SBW8 A0A315W2H7 A0A1A8VAG0 A0A3Q2D080 A0A182J1F2 A0A3B4UFP1 A0A1A6HRT1 Q32KY0 A0A3Q0QX35 A0A182LEN1 F1MS32 A0A3P8U4T7 F5HKU4 A0A0K8UND9 L8IIL2 A0A182XUT6 A0A2C9GRE4 A0A1S3FFP1 D2HH45 G1LS67 C3YC60 F6XM13 P51910 A0A3Q0CSM2 A0A340XXN3 G3T8L4 A0A1D2MJS8 P23593 A0A3B5MND9 M0R4S2 A0A1D2MVQ6
Pubmed
19121390
28756777
23622113
22118469
26354079
15496914
+ More
17554307 24438588 23542700 29451363 26227816 24487278 1572353 25186727 6386809 3608987 4096898 21551351 23258410 24495485 29703783 30723633 20966253 19393038 12364791 14747013 17210077 22751099 20010809 18563158 19892987 7637575 8666283 16141072 15489334 21183079 26319212 27289101 1695148 2090718 15057822 15632090
17554307 24438588 23542700 29451363 26227816 24487278 1572353 25186727 6386809 3608987 4096898 21551351 23258410 24495485 29703783 30723633 20966253 19393038 12364791 14747013 17210077 22751099 20010809 18563158 19892987 7637575 8666283 16141072 15489334 21183079 26319212 27289101 1695148 2090718 15057822 15632090
EMBL
BABH01025083
BABH01025084
ODYU01003843
SOQ43103.1
KZ149918
PZC77809.1
+ More
GAIX01008028 JAA84532.1 NWSH01000611 PCG75316.1 AGBW02014563 OWR41413.1 KQ460205 KPJ17086.1 PCG75317.1 KQ459604 KPI92164.1 CAAE01014568 CAF98955.1 KPI92165.1 ATLV01026880 KE525420 KFB53445.1 CP026249 AWP03818.1 GCES01005579 JAR80744.1 AYCK01009881 SOQ43104.1 JTDY01004480 KOB68097.1 PZC77810.1 X64714 CAA45969.1 KPJ17087.1 PCG75315.1 AERX01019766 GDQN01005045 GDQN01003677 JAT86009.1 JAT87377.1 L41641 AAA85089.1 KU096845 AMY63178.1 GBYX01069461 JAO99769.1 X64715 AB294515 BAF47403.1 KB030403 ELK17614.1 GAMC01002289 JAC04267.1 NHOQ01000466 PWA29964.1 AEMK02000089 DQIR01310312 HDC65790.1 PWA30007.1 HAEJ01016135 SBS56592.1 LZPO01017321 OBS80720.1 BC109863 AAAB01008859 EGK96846.1 GDHF01024208 JAI28106.1 JH881342 ELR54927.1 APCN01000283 GL192829 EFB14744.1 ACTA01050789 GG666500 EEN62092.1 L39123 X82648 AK135046 AK157917 AK158118 AK158405 AK160729 AK162392 BC145907 BC145909 LJIJ01001041 ODM93243.1 X55572 AABR07034459 CH473967 EDM11470.1 LJIJ01000467 ODM97170.1
GAIX01008028 JAA84532.1 NWSH01000611 PCG75316.1 AGBW02014563 OWR41413.1 KQ460205 KPJ17086.1 PCG75317.1 KQ459604 KPI92164.1 CAAE01014568 CAF98955.1 KPI92165.1 ATLV01026880 KE525420 KFB53445.1 CP026249 AWP03818.1 GCES01005579 JAR80744.1 AYCK01009881 SOQ43104.1 JTDY01004480 KOB68097.1 PZC77810.1 X64714 CAA45969.1 KPJ17087.1 PCG75315.1 AERX01019766 GDQN01005045 GDQN01003677 JAT86009.1 JAT87377.1 L41641 AAA85089.1 KU096845 AMY63178.1 GBYX01069461 JAO99769.1 X64715 AB294515 BAF47403.1 KB030403 ELK17614.1 GAMC01002289 JAC04267.1 NHOQ01000466 PWA29964.1 AEMK02000089 DQIR01310312 HDC65790.1 PWA30007.1 HAEJ01016135 SBS56592.1 LZPO01017321 OBS80720.1 BC109863 AAAB01008859 EGK96846.1 GDHF01024208 JAI28106.1 JH881342 ELR54927.1 APCN01000283 GL192829 EFB14744.1 ACTA01050789 GG666500 EEN62092.1 L39123 X82648 AK135046 AK157917 AK158118 AK158405 AK160729 AK162392 BC145907 BC145909 LJIJ01001041 ODM93243.1 X55572 AABR07034459 CH473967 EDM11470.1 LJIJ01000467 ODM97170.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053240
UP000053268
UP000001038
+ More
UP000265180 UP000265200 UP000030765 UP000002852 UP000246464 UP000261560 UP000261420 UP000028760 UP000261500 UP000265000 UP000037510 UP000242638 UP000265120 UP000261480 UP000257200 UP000261340 UP000005207 UP000264840 UP000265160 UP000265100 UP000261580 UP000007635 UP000261620 UP000005226 UP000265040 UP000261460 UP000261400 UP000265020 UP000010552 UP000008227 UP000257160 UP000075880 UP000092124 UP000009136 UP000075882 UP000265080 UP000007062 UP000076407 UP000075840 UP000081671 UP000008912 UP000001554 UP000002281 UP000000589 UP000189706 UP000265300 UP000007646 UP000094527 UP000002494 UP000261380
UP000265180 UP000265200 UP000030765 UP000002852 UP000246464 UP000261560 UP000261420 UP000028760 UP000261500 UP000265000 UP000037510 UP000242638 UP000265120 UP000261480 UP000257200 UP000261340 UP000005207 UP000264840 UP000265160 UP000265100 UP000261580 UP000007635 UP000261620 UP000005226 UP000265040 UP000261460 UP000261400 UP000265020 UP000010552 UP000008227 UP000257160 UP000075880 UP000092124 UP000009136 UP000075882 UP000265080 UP000007062 UP000076407 UP000075840 UP000081671 UP000008912 UP000001554 UP000002281 UP000000589 UP000189706 UP000265300 UP000007646 UP000094527 UP000002494 UP000261380
Interpro
SUPFAM
SSF50814
SSF50814
Gene 3D
ProteinModelPortal
H9J5S6
A0A2H1VQH8
A0A2W1BRV6
S4PVS5
A0A2A4JU62
A0A212EIU3
+ More
A0A194RM77 A0A2A4JTF5 A0A194PHH4 Q4SK77 H2MUK0 H2MUJ9 A0A3P9K5L3 A0A3P9K5M3 A0A3P9IV14 A0A3P9H6J3 A0A194PM29 A0A084WTA1 M4ATP2 A0A2U9BID1 A0A3B3DF18 A0A147AQ88 A0A3B4UH23 A0A087XJ49 A0A3B3U536 A0A3Q2P803 A0A2H1VSA5 A0A0L7KXZ8 A0A2W1BYB5 A0A3P9PEY2 A0A3P8W739 A0A3B3X9V5 A0A3Q1HHN3 Q00629 A0A194RHP6 A0A3Q0RF77 A0A3Q0RKG1 A0A2A4JTE9 I3KQQ7 A0A3Q1HDT0 A0A1E1WGA5 A0A3Q2VUM2 A0A3P9DDR8 P00305 Q24996 A0A3P8PHW7 A0A3Q4HDD4 G3PXV7 A0A3Q4BXK7 A0A159YQU9 A0A3Q3XJW8 H2SCU4 A0A0S7MDZ3 Q00630 A0A3Q1JNI9 A0A3B4FLM3 A3KEW8 A0A3B5A5P2 A0A3Q2D0T8 L5L1J2 W8C9C1 A0A315W1Y7 A0A3Q1KCJ4 G3PXW5 F1SQX9 A0A3Q1CF05 H2SBW8 A0A315W2H7 A0A1A8VAG0 A0A3Q2D080 A0A182J1F2 A0A3B4UFP1 A0A1A6HRT1 Q32KY0 A0A3Q0QX35 A0A182LEN1 F1MS32 A0A3P8U4T7 F5HKU4 A0A0K8UND9 L8IIL2 A0A182XUT6 A0A2C9GRE4 A0A1S3FFP1 D2HH45 G1LS67 C3YC60 F6XM13 P51910 A0A3Q0CSM2 A0A340XXN3 G3T8L4 A0A1D2MJS8 P23593 A0A3B5MND9 M0R4S2 A0A1D2MVQ6
A0A194RM77 A0A2A4JTF5 A0A194PHH4 Q4SK77 H2MUK0 H2MUJ9 A0A3P9K5L3 A0A3P9K5M3 A0A3P9IV14 A0A3P9H6J3 A0A194PM29 A0A084WTA1 M4ATP2 A0A2U9BID1 A0A3B3DF18 A0A147AQ88 A0A3B4UH23 A0A087XJ49 A0A3B3U536 A0A3Q2P803 A0A2H1VSA5 A0A0L7KXZ8 A0A2W1BYB5 A0A3P9PEY2 A0A3P8W739 A0A3B3X9V5 A0A3Q1HHN3 Q00629 A0A194RHP6 A0A3Q0RF77 A0A3Q0RKG1 A0A2A4JTE9 I3KQQ7 A0A3Q1HDT0 A0A1E1WGA5 A0A3Q2VUM2 A0A3P9DDR8 P00305 Q24996 A0A3P8PHW7 A0A3Q4HDD4 G3PXV7 A0A3Q4BXK7 A0A159YQU9 A0A3Q3XJW8 H2SCU4 A0A0S7MDZ3 Q00630 A0A3Q1JNI9 A0A3B4FLM3 A3KEW8 A0A3B5A5P2 A0A3Q2D0T8 L5L1J2 W8C9C1 A0A315W1Y7 A0A3Q1KCJ4 G3PXW5 F1SQX9 A0A3Q1CF05 H2SBW8 A0A315W2H7 A0A1A8VAG0 A0A3Q2D080 A0A182J1F2 A0A3B4UFP1 A0A1A6HRT1 Q32KY0 A0A3Q0QX35 A0A182LEN1 F1MS32 A0A3P8U4T7 F5HKU4 A0A0K8UND9 L8IIL2 A0A182XUT6 A0A2C9GRE4 A0A1S3FFP1 D2HH45 G1LS67 C3YC60 F6XM13 P51910 A0A3Q0CSM2 A0A340XXN3 G3T8L4 A0A1D2MJS8 P23593 A0A3B5MND9 M0R4S2 A0A1D2MVQ6
PDB
1Z24
E-value=8.86978e-12,
Score=166
Ontologies
GO
GO:0031409
GO:0006869
GO:0008289
GO:0007568
GO:0000302
GO:0006629
GO:0005737
GO:0034632
GO:0005501
GO:0018298
GO:0005576
GO:0010642
GO:0048678
GO:0048471
GO:0042493
GO:0042246
GO:2000098
GO:0071638
GO:0005615
GO:2000405
GO:0060588
GO:0005783
GO:0030425
GO:1900016
GO:0042308
GO:0022626
GO:0048662
GO:0051895
GO:0014012
GO:0006006
GO:0015485
GO:0007420
GO:0043025
GO:0006508
GO:0008270
GO:0004180
GO:0005515
GO:0003676
GO:0007154
GO:0016020
GO:0005112
GO:0015930
Topology
Subcellular location
Secreted
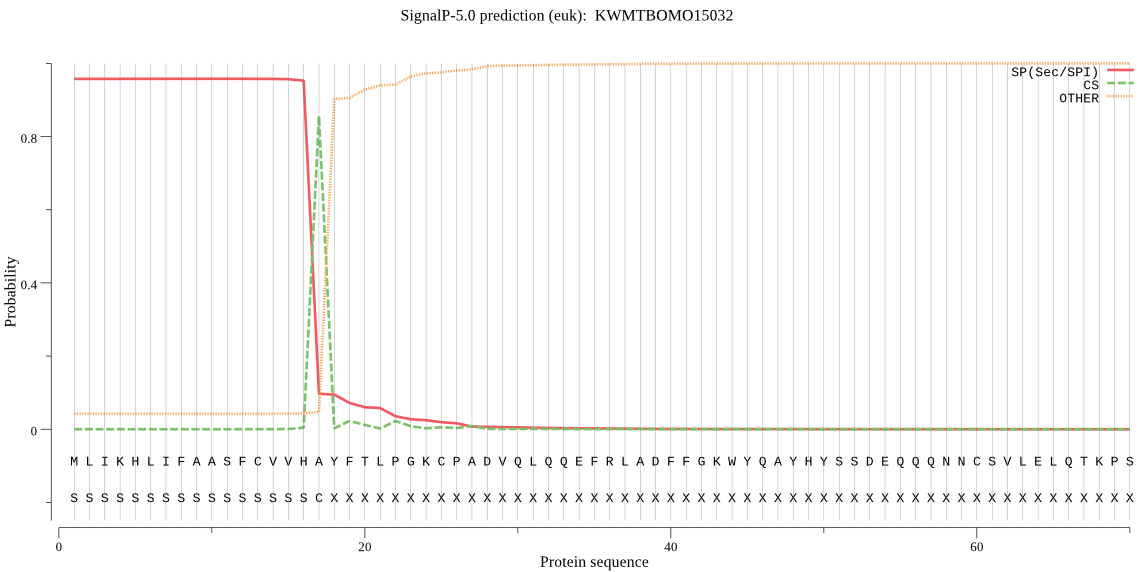
SignalP
Position: 1 - 17,
Likelihood: 0.957438
Length:
213
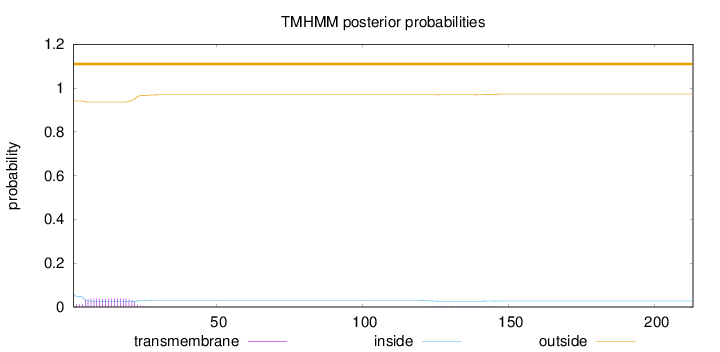
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.81505
Exp number, first 60 AAs:
0.71892
Total prob of N-in:
0.05901
outside
1 - 213
Population Genetic Test Statistics
Pi
136.503875
Theta
139.923783
Tajima's D
0.052305
CLR
0.553966
CSRT
0.380830958452077
Interpretation
Uncertain
