Gene
KWMTBOMO15024
Pre Gene Modal
BGIBMGA004801
Annotation
PREDICTED:_LOW_QUALITY_PROTEIN:_uncharacterized_protein_LOC101745803_[Bombyx_mori]
Full name
Zinc carboxypeptidase
Location in the cell
PlasmaMembrane Reliability : 2.691
Sequence
CDS
ATGCAATGGGATGCGTATTATGAACTGGAAGAAATATACGCCTGGATAGATGATTTAGCAAAAACATATCCAGATATTGTCACAACAGAAGTCGCTGGTACTTCATATGAAGGTCGAGAAATTAAGGGATTGAAAATATCCCATGGGCCCGGTAGAAGGATAGTGTTTATCGAAAGCGGTATTCATGCTAGAGAATGGATCACTCCAGCTACAGTATGCTATATCACCGCCCAGTTGCTTACTAGTAACGATCCAGAGACAAGAGCAGCAGCAAGAGATTTCGACTGGTATATATTCCCTGTGACGAACCCAGATGGGTATGTTTGGACACACACTGATTTTCGGCTGTGGCGCAAAAACAGAAAGCCAGTCGGTACAGAATTCGGTATTGACTTAAATAGAAACTGGAACAGTAACTGGTTGGTTAAAGGCTCCAGTACTAACCCGTCTTCAAATAACTACGCTGGTCTTGGACCATTCTCGGAACCCGAGACCCGCACTCTTTCCACATACATAACGGCTATCGGAAATAAAATTGATATATACCTGTCTATGCATTCCTCTGGCCAGCTCTTATTGGTACCTTTTGGTAACACAACTCAGCCTTTGGCAAATTATTATGATGTGGTGAATATTGGAAGGCGTGCGATGGGAGCTTTATCAGTGAGATACGGTACAGAATATACAACAGGAAATGTAGCTGAAGCCATATACCATGCAACAGGAAGCAGCGCAGATTGGGTCAAAGAGTTCCTTGACGTCCCCTTGGTCTACATATACGAGCTCCGTGACAGGGGAACTTTCGGTCATCTCCTGCCTCCTGATCAGATCTTGCCCACCAGCGAAGAGACAATGGACTCTATAATCGACCTCATCCACCAAGCCAAGAGATTCGGGTACATGAACAGCGGAAACGGACTAAAATTTTCATTTTTGACTCCACTACTGCTTTTCGTTGTTAAATCTGTTGTGTGA
Protein
MQWDAYYELEEIYAWIDDLAKTYPDIVTTEVAGTSYEGREIKGLKISHGPGRRIVFIESGIHAREWITPATVCYITAQLLTSNDPETRAAARDFDWYIFPVTNPDGYVWTHTDFRLWRKNRKPVGTEFGIDLNRNWNSNWLVKGSSTNPSSNNYAGLGPFSEPETRTLSTYITAIGNKIDIYLSMHSSGQLLLVPFGNTTQPLANYYDVVNIGRRAMGALSVRYGTEYTTGNVAEAIYHATGSSADWVKEFLDVPLVYIYELRDRGTFGHLLPPDQILPTSEETMDSIIDLIHQAKRFGYMNSGNGLKFSFLTPLLLFVVKSVV
Summary
Cofactor
Zn(2+)
Similarity
Belongs to the peptidase M14 family.
Keywords
Carboxypeptidase
Disulfide bond
Hydrolase
Metal-binding
Metalloprotease
Protease
Secreted
Zinc
Feature
chain Zinc carboxypeptidase
Uniprot
A0A2W1BNR4
A0A2A4JQ49
H9J5L1
A0A1E1WDG3
A0A2H1X3V0
A0A2W1BNU0
+ More
A0A194PHG9 A0A3S2LHE6 A0A0L7KXG1 A0A194RIC8 A0A182H3R1 A0A3G5BIF5 A0A2A3E2U1 A0A1S4F4N5 Q6J6D5 A0A0C9RFV3 A0A195FPR9 W5JN76 A0A084VIH2 A0A195CUQ5 A0A088A4S2 A0A182TX13 A0A182QY48 A0A182V379 A0A195FPQ9 Q7Q316 A0A195CTS9 A0A195BNW8 A0A0J7KVA1 A0A158NTV3 B0WS11 A0A182LXL8 A0A1I8JW63 A0A182LPK9 A0A2C9GR29 A0A158P468 K7IN13 E9INB0 A0A154PKT4 A0A1B0DP13 F4MI68 A0A026WMA6 E1ZXR6 A0A2P8YM67 F4WJ50 A0A195FPG7 A0A310SEP0 A0A1L8DR93 F4WIQ2 A0A1L8DQV7 Q5TPZ3 A0A182UYU3 B0WS12 E2BM37 A0A232EGG7 W5JK64 V9QNN0 K7IY61 A0A195BNH2 A0A182QAV5 A0A151JRL5 A0A182K4R6 A0A151WLL6 E1ZXR7 A0A182TPH8 A0A0M8ZXY7 A0A1I8JW39 A0A182P9F0 A0A182LPL5 A0A2C9GQU4 Q17FG0 A0A182RFC6 A0A182LT40 A0A182JEM2 A0A232F1J1 A0A310SCR6 A0A182FUU7 A0A182MXY6 F4WMS6 P42788 A0A2J7REK7 A0A154PJT5 A0A182WF36 A0A182YDB7 A0A158NQ85 F4WMS7 U5EWN0 A0A195CTW9 A0A158NQ44 A0A195FPB2 A0A1L8DR23 A0A067RGG5 A0A194PGK0 A0A336KG85 A0A2J7REM7 A8CW74 A0A1V1FIW8 A0A1B0EV43 A8CAE9 A0A336MR26
A0A194PHG9 A0A3S2LHE6 A0A0L7KXG1 A0A194RIC8 A0A182H3R1 A0A3G5BIF5 A0A2A3E2U1 A0A1S4F4N5 Q6J6D5 A0A0C9RFV3 A0A195FPR9 W5JN76 A0A084VIH2 A0A195CUQ5 A0A088A4S2 A0A182TX13 A0A182QY48 A0A182V379 A0A195FPQ9 Q7Q316 A0A195CTS9 A0A195BNW8 A0A0J7KVA1 A0A158NTV3 B0WS11 A0A182LXL8 A0A1I8JW63 A0A182LPK9 A0A2C9GR29 A0A158P468 K7IN13 E9INB0 A0A154PKT4 A0A1B0DP13 F4MI68 A0A026WMA6 E1ZXR6 A0A2P8YM67 F4WJ50 A0A195FPG7 A0A310SEP0 A0A1L8DR93 F4WIQ2 A0A1L8DQV7 Q5TPZ3 A0A182UYU3 B0WS12 E2BM37 A0A232EGG7 W5JK64 V9QNN0 K7IY61 A0A195BNH2 A0A182QAV5 A0A151JRL5 A0A182K4R6 A0A151WLL6 E1ZXR7 A0A182TPH8 A0A0M8ZXY7 A0A1I8JW39 A0A182P9F0 A0A182LPL5 A0A2C9GQU4 Q17FG0 A0A182RFC6 A0A182LT40 A0A182JEM2 A0A232F1J1 A0A310SCR6 A0A182FUU7 A0A182MXY6 F4WMS6 P42788 A0A2J7REK7 A0A154PJT5 A0A182WF36 A0A182YDB7 A0A158NQ85 F4WMS7 U5EWN0 A0A195CTW9 A0A158NQ44 A0A195FPB2 A0A1L8DR23 A0A067RGG5 A0A194PGK0 A0A336KG85 A0A2J7REM7 A8CW74 A0A1V1FIW8 A0A1B0EV43 A8CAE9 A0A336MR26
EC Number
3.4.17.-
Pubmed
EMBL
KZ150003
PZC75274.1
NWSH01000796
PCG74167.1
BABH01025123
BABH01025124
+ More
GDQN01007227 GDQN01006034 JAT83827.1 JAT85020.1 ODYU01013306 SOQ60013.1 PZC75275.1 KQ459604 KPI92159.1 RSAL01000009 RVE53889.1 JTDY01004606 KOB67958.1 KQ460205 KPJ17080.1 JXUM01024706 KQ560699 KXJ81228.1 MK075163 AYV99566.1 KZ288417 PBC26020.1 AY590492 AAT36730.1 GBYB01012027 JAG81794.1 KQ981382 KYN42302.1 ADMH02000883 ETN64768.1 ATLV01013356 KE524854 KFB37766.1 KQ977279 KYN04252.1 AXCN02002112 KYN42292.1 AAAB01008966 EAA13019.4 KYN04096.1 KQ976435 KYM87054.1 LBMM01002732 KMQ94422.1 ADTU01026166 DS232063 EDS33629.1 AXCM01005733 APCN01001947 ADTU01008675 GL764292 EFZ17948.1 KQ434938 KZC12084.1 AJVK01017975 EZ966131 ADL13888.1 KK107167 QOIP01000010 EZA56244.1 RLU17286.1 GL435087 EFN74038.1 PYGN01000496 PSN45347.1 GL888181 EGI65820.1 KYN42291.1 KQ769940 OAD52755.1 GFDF01005229 JAV08855.1 GL888177 EGI65848.1 GFDF01005228 JAV08856.1 EAL39491.3 EDS33630.1 GL449129 EFN83243.1 NNAY01004759 OXU17455.1 ETN64767.1 KF707629 AHC32031.1 KFB37767.1 KYM87041.1 KQ978615 KYN29840.1 KQ982959 KYQ48754.1 EFN74039.1 KQ435828 KOX71863.1 CH477271 EAT45285.1 NNAY01001310 OXU24399.1 OAD52756.1 GL888218 EGI64553.1 L08481 NEVH01004959 PNF39274.1 KZC12083.1 ADTU01022957 EGI64554.1 GANO01002887 JAB56984.1 KYN04105.1 KYN42152.1 GFDF01005233 JAV08851.1 KK852482 KDR22872.1 KPI92158.1 UFQS01000434 UFQT01000434 SSX03896.1 SSX24261.1 PNF39276.1 EU124592 ABV60310.1 FX985401 BAX07414.1 AJWK01015367 EU045341 ABV44738.1 UFQT01001473 SSX30717.1
GDQN01007227 GDQN01006034 JAT83827.1 JAT85020.1 ODYU01013306 SOQ60013.1 PZC75275.1 KQ459604 KPI92159.1 RSAL01000009 RVE53889.1 JTDY01004606 KOB67958.1 KQ460205 KPJ17080.1 JXUM01024706 KQ560699 KXJ81228.1 MK075163 AYV99566.1 KZ288417 PBC26020.1 AY590492 AAT36730.1 GBYB01012027 JAG81794.1 KQ981382 KYN42302.1 ADMH02000883 ETN64768.1 ATLV01013356 KE524854 KFB37766.1 KQ977279 KYN04252.1 AXCN02002112 KYN42292.1 AAAB01008966 EAA13019.4 KYN04096.1 KQ976435 KYM87054.1 LBMM01002732 KMQ94422.1 ADTU01026166 DS232063 EDS33629.1 AXCM01005733 APCN01001947 ADTU01008675 GL764292 EFZ17948.1 KQ434938 KZC12084.1 AJVK01017975 EZ966131 ADL13888.1 KK107167 QOIP01000010 EZA56244.1 RLU17286.1 GL435087 EFN74038.1 PYGN01000496 PSN45347.1 GL888181 EGI65820.1 KYN42291.1 KQ769940 OAD52755.1 GFDF01005229 JAV08855.1 GL888177 EGI65848.1 GFDF01005228 JAV08856.1 EAL39491.3 EDS33630.1 GL449129 EFN83243.1 NNAY01004759 OXU17455.1 ETN64767.1 KF707629 AHC32031.1 KFB37767.1 KYM87041.1 KQ978615 KYN29840.1 KQ982959 KYQ48754.1 EFN74039.1 KQ435828 KOX71863.1 CH477271 EAT45285.1 NNAY01001310 OXU24399.1 OAD52756.1 GL888218 EGI64553.1 L08481 NEVH01004959 PNF39274.1 KZC12083.1 ADTU01022957 EGI64554.1 GANO01002887 JAB56984.1 KYN04105.1 KYN42152.1 GFDF01005233 JAV08851.1 KK852482 KDR22872.1 KPI92158.1 UFQS01000434 UFQT01000434 SSX03896.1 SSX24261.1 PNF39276.1 EU124592 ABV60310.1 FX985401 BAX07414.1 AJWK01015367 EU045341 ABV44738.1 UFQT01001473 SSX30717.1
Proteomes
UP000218220
UP000005204
UP000053268
UP000283053
UP000037510
UP000053240
+ More
UP000069940 UP000249989 UP000242457 UP000078541 UP000000673 UP000030765 UP000078542 UP000005203 UP000075902 UP000075886 UP000075903 UP000007062 UP000078540 UP000036403 UP000005205 UP000002320 UP000075883 UP000076407 UP000075882 UP000075840 UP000002358 UP000076502 UP000092462 UP000053097 UP000279307 UP000000311 UP000245037 UP000007755 UP000008237 UP000215335 UP000078492 UP000075881 UP000075809 UP000053105 UP000075885 UP000008820 UP000075900 UP000075880 UP000069272 UP000075884 UP000235965 UP000075920 UP000076408 UP000027135 UP000092461
UP000069940 UP000249989 UP000242457 UP000078541 UP000000673 UP000030765 UP000078542 UP000005203 UP000075902 UP000075886 UP000075903 UP000007062 UP000078540 UP000036403 UP000005205 UP000002320 UP000075883 UP000076407 UP000075882 UP000075840 UP000002358 UP000076502 UP000092462 UP000053097 UP000279307 UP000000311 UP000245037 UP000007755 UP000008237 UP000215335 UP000078492 UP000075881 UP000075809 UP000053105 UP000075885 UP000008820 UP000075900 UP000075880 UP000069272 UP000075884 UP000235965 UP000075920 UP000076408 UP000027135 UP000092461
PRIDE
Interpro
Gene 3D
ProteinModelPortal
A0A2W1BNR4
A0A2A4JQ49
H9J5L1
A0A1E1WDG3
A0A2H1X3V0
A0A2W1BNU0
+ More
A0A194PHG9 A0A3S2LHE6 A0A0L7KXG1 A0A194RIC8 A0A182H3R1 A0A3G5BIF5 A0A2A3E2U1 A0A1S4F4N5 Q6J6D5 A0A0C9RFV3 A0A195FPR9 W5JN76 A0A084VIH2 A0A195CUQ5 A0A088A4S2 A0A182TX13 A0A182QY48 A0A182V379 A0A195FPQ9 Q7Q316 A0A195CTS9 A0A195BNW8 A0A0J7KVA1 A0A158NTV3 B0WS11 A0A182LXL8 A0A1I8JW63 A0A182LPK9 A0A2C9GR29 A0A158P468 K7IN13 E9INB0 A0A154PKT4 A0A1B0DP13 F4MI68 A0A026WMA6 E1ZXR6 A0A2P8YM67 F4WJ50 A0A195FPG7 A0A310SEP0 A0A1L8DR93 F4WIQ2 A0A1L8DQV7 Q5TPZ3 A0A182UYU3 B0WS12 E2BM37 A0A232EGG7 W5JK64 V9QNN0 K7IY61 A0A195BNH2 A0A182QAV5 A0A151JRL5 A0A182K4R6 A0A151WLL6 E1ZXR7 A0A182TPH8 A0A0M8ZXY7 A0A1I8JW39 A0A182P9F0 A0A182LPL5 A0A2C9GQU4 Q17FG0 A0A182RFC6 A0A182LT40 A0A182JEM2 A0A232F1J1 A0A310SCR6 A0A182FUU7 A0A182MXY6 F4WMS6 P42788 A0A2J7REK7 A0A154PJT5 A0A182WF36 A0A182YDB7 A0A158NQ85 F4WMS7 U5EWN0 A0A195CTW9 A0A158NQ44 A0A195FPB2 A0A1L8DR23 A0A067RGG5 A0A194PGK0 A0A336KG85 A0A2J7REM7 A8CW74 A0A1V1FIW8 A0A1B0EV43 A8CAE9 A0A336MR26
A0A194PHG9 A0A3S2LHE6 A0A0L7KXG1 A0A194RIC8 A0A182H3R1 A0A3G5BIF5 A0A2A3E2U1 A0A1S4F4N5 Q6J6D5 A0A0C9RFV3 A0A195FPR9 W5JN76 A0A084VIH2 A0A195CUQ5 A0A088A4S2 A0A182TX13 A0A182QY48 A0A182V379 A0A195FPQ9 Q7Q316 A0A195CTS9 A0A195BNW8 A0A0J7KVA1 A0A158NTV3 B0WS11 A0A182LXL8 A0A1I8JW63 A0A182LPK9 A0A2C9GR29 A0A158P468 K7IN13 E9INB0 A0A154PKT4 A0A1B0DP13 F4MI68 A0A026WMA6 E1ZXR6 A0A2P8YM67 F4WJ50 A0A195FPG7 A0A310SEP0 A0A1L8DR93 F4WIQ2 A0A1L8DQV7 Q5TPZ3 A0A182UYU3 B0WS12 E2BM37 A0A232EGG7 W5JK64 V9QNN0 K7IY61 A0A195BNH2 A0A182QAV5 A0A151JRL5 A0A182K4R6 A0A151WLL6 E1ZXR7 A0A182TPH8 A0A0M8ZXY7 A0A1I8JW39 A0A182P9F0 A0A182LPL5 A0A2C9GQU4 Q17FG0 A0A182RFC6 A0A182LT40 A0A182JEM2 A0A232F1J1 A0A310SCR6 A0A182FUU7 A0A182MXY6 F4WMS6 P42788 A0A2J7REK7 A0A154PJT5 A0A182WF36 A0A182YDB7 A0A158NQ85 F4WMS7 U5EWN0 A0A195CTW9 A0A158NQ44 A0A195FPB2 A0A1L8DR23 A0A067RGG5 A0A194PGK0 A0A336KG85 A0A2J7REM7 A8CW74 A0A1V1FIW8 A0A1B0EV43 A8CAE9 A0A336MR26
PDB
5MRV
E-value=1.21578e-65,
Score=633
Ontologies
GO
Topology
Subcellular location
Secreted
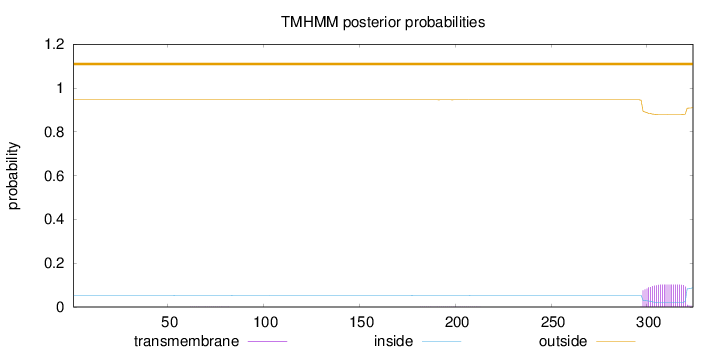
Length:
324
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
2.27569
Exp number, first 60 AAs:
0.00012
Total prob of N-in:
0.05333
outside
1 - 324
Population Genetic Test Statistics
Pi
163.984119
Theta
142.695426
Tajima's D
0
CLR
0.512557
CSRT
0.362781860906955
Interpretation
Uncertain
