Gene
KWMTBOMO15022
Annotation
PREDICTED:_mast_cell_carboxypeptidase_A-like_[Bombyx_mori]
Full name
Mast cell carboxypeptidase A
Alternative Name
Carboxypeptidase A3
Location in the cell
PlasmaMembrane Reliability : 0.989
Sequence
CDS
ATGTCTAATTTCGTGAGAAACCACAGTAACCATCTGGAATACTATTTGGCCTTCCATTCGTACGGGCAAGCCATGATCCTGCCTTACGCGAATACAAGAGCTCACATGGAAAATTTTGATACTGTGGCAGCCATGGCTAAACAAGCAGCAGACCACATAGCTCTCAGATACAATACCAGATACGTGTATGGAACAGCTTATGACACTGTAGGTTACGTGACATCCGGCGTCAGTGGTTGCTGGGTCAAAGAAACATTTAGTGTTCCGTACGTCCTTACATTTGAACTTCCCGACCGAGGGCAATATGGTTTCGCTCTGCCCAATGAGAAGATCACGCCAACATGTATAGAAACCATGGATGGTGTTTTGTCACTATTGAAACCTCGCACGAAAATTTACGATAAACTAAGACACGAGCACAGGGGTTTCGATGACGGTCAAAGGACGAAGTTTTTGGATAAATTGGTGATCCTTTTTAATGTGTGCCTGTTTGTTATTGCTCGTTTTGATTAA
Protein
MSNFVRNHSNHLEYYLAFHSYGQAMILPYANTRAHMENFDTVAAMAKQAADHIALRYNTRYVYGTAYDTVGYVTSGVSGCWVKETFSVPYVLTFELPDRGQYGFALPNEKITPTCIETMDGVLSLLKPRTKIYDKLRHEHRGFDDGQRTKFLDKLVILFNVCLFVIARFD
Summary
Catalytic Activity
Release of a C-terminal amino acid, but little or no action with -Asp, -Glu, -Arg, -Lys or -Pro.
Cofactor
Zn(2+)
Similarity
Belongs to the peptidase M14 family.
Keywords
Carboxypeptidase
Complete proteome
Cytoplasmic vesicle
Direct protein sequencing
Disulfide bond
Hydrolase
Metal-binding
Metalloprotease
Polymorphism
Protease
Reference proteome
Signal
Zinc
Zymogen
Feature
propeptide Activation peptide
chain Mast cell carboxypeptidase A
sequence variant In dbSNP:rs2270523.
chain Mast cell carboxypeptidase A
sequence variant In dbSNP:rs2270523.
Uniprot
A0A194RGY2
A0A212F6I3
A0A2A4J3C4
A0A2W1BJL8
K7IN13
A0A310SEP0
+ More
A0A088A4S2 V5G180 A0A2A3E2U1 A0A067RGG5 V5I8H1 V5I8J9 A0A154PKT4 A0A182LX23 B0WS12 A0A0L7R9B9 A0A195BNW8 A0A195CUQ5 A0A158NTV3 A0A2K6V6C3 A0A2K5PME4 A0A1V1FIW8 A0A0C9RFV3 A0A158NQ44 A0A2A4JQ49 A0A195CTS9 D2A3Y2 N6T3L7 U4UC85 K7IQY4 A0A182P9F1 F4WMS6 F7HXH8 A0A2K5EUS2 A0A182YDB7 A0A0M8ZXY7 A0A1L8DR93 A0A1L8DQV7 A1XGA1 A0A182LT40 A1XGA0 A1XG97 H2PBP8 A0A0L7R992 A0A2W1BNU0 K7JA32 A1XG98 A0A195BHV6 A0A232EGG7 A0A232F1J1 K7IY61 L5JVM8 A0A182QAV5 A0A310SCR6 A0A0J7KVA1 A0A2I3TU16 A1XGA3 A0A2R9CAY4 A0A3Q0D5I3 A0A096P028 A0A0L7KXG1 H2QNJ6 A0A195FPB2 S7MU95 A0A2K6KUE9 A0A2R9CLS4 A0A2J8P2V4 B2R941 P15088 A0A158NQ85 A1XGA2 A0A2K6C4D9 G7MJJ7 H9J5L1 G7NZN8 F7GFE7 A0A2K5WCY7 Q542E3 P15089 G3S533 A0A2K6LW11 A0A2K6LVY0 A0A0D9RG95 A0A2K5JGG1 A0A3G5BIF5 A0A2K5CNU8 A0A151WLL6 L5MAR4 A0A151J021 A0A2S2N6Y1 A0A2K5R360 A0A2K5CNV4 A0A341B9Q3 A0A2K6NNH6
A0A088A4S2 V5G180 A0A2A3E2U1 A0A067RGG5 V5I8H1 V5I8J9 A0A154PKT4 A0A182LX23 B0WS12 A0A0L7R9B9 A0A195BNW8 A0A195CUQ5 A0A158NTV3 A0A2K6V6C3 A0A2K5PME4 A0A1V1FIW8 A0A0C9RFV3 A0A158NQ44 A0A2A4JQ49 A0A195CTS9 D2A3Y2 N6T3L7 U4UC85 K7IQY4 A0A182P9F1 F4WMS6 F7HXH8 A0A2K5EUS2 A0A182YDB7 A0A0M8ZXY7 A0A1L8DR93 A0A1L8DQV7 A1XGA1 A0A182LT40 A1XGA0 A1XG97 H2PBP8 A0A0L7R992 A0A2W1BNU0 K7JA32 A1XG98 A0A195BHV6 A0A232EGG7 A0A232F1J1 K7IY61 L5JVM8 A0A182QAV5 A0A310SCR6 A0A0J7KVA1 A0A2I3TU16 A1XGA3 A0A2R9CAY4 A0A3Q0D5I3 A0A096P028 A0A0L7KXG1 H2QNJ6 A0A195FPB2 S7MU95 A0A2K6KUE9 A0A2R9CLS4 A0A2J8P2V4 B2R941 P15088 A0A158NQ85 A1XGA2 A0A2K6C4D9 G7MJJ7 H9J5L1 G7NZN8 F7GFE7 A0A2K5WCY7 Q542E3 P15089 G3S533 A0A2K6LW11 A0A2K6LVY0 A0A0D9RG95 A0A2K5JGG1 A0A3G5BIF5 A0A2K5CNU8 A0A151WLL6 L5MAR4 A0A151J021 A0A2S2N6Y1 A0A2K5R360 A0A2K5CNV4 A0A341B9Q3 A0A2K6NNH6
EC Number
3.4.17.1
Pubmed
26354079
22118469
28756777
20075255
24845553
21347285
+ More
28410430 18362917 19820115 23537049 21719571 25244985 17651235 28648823 23258410 16136131 22722832 26319212 26227816 2594780 1729276 16641997 15489334 1629626 2708524 22002653 19121390 17431167 10349636 11042159 11076861 11217851 12466851 12040188 16141073 2584208 22398555 30400621 25362486
28410430 18362917 19820115 23537049 21719571 25244985 17651235 28648823 23258410 16136131 22722832 26319212 26227816 2594780 1729276 16641997 15489334 1629626 2708524 22002653 19121390 17431167 10349636 11042159 11076861 11217851 12466851 12040188 16141073 2584208 22398555 30400621 25362486
EMBL
KQ460205
KPJ17078.1
AGBW02010033
OWR49319.1
NWSH01003708
PCG65902.1
+ More
KZ150003 PZC75272.1 KQ769940 OAD52755.1 GALX01004696 JAB63770.1 KZ288417 PBC26020.1 KK852482 KDR22872.1 GALX01004695 JAB63771.1 GALX01004555 JAB63911.1 KQ434938 KZC12084.1 AXCM01002791 DS232063 EDS33630.1 KQ414627 KOC67443.1 KQ976435 KYM87054.1 KQ977279 KYN04252.1 ADTU01026166 FX985401 BAX07414.1 GBYB01012027 JAG81794.1 ADTU01022957 NWSH01000796 PCG74167.1 KYN04096.1 KQ971348 EFA05749.1 APGK01045325 APGK01045326 KB741033 ENN74749.1 KB631976 ERL87560.1 GL888218 EGI64553.1 KQ435828 KOX71863.1 GFDF01005229 JAV08855.1 GFDF01005228 JAV08856.1 DQ356060 ABC88775.1 AXCM01005733 DQ356059 ABC88774.1 DQ356056 ABC88771.1 ABGA01105389 ABGA01105390 NDHI03003379 PNJ72804.1 KOC67445.1 PZC75275.1 AAZX01005059 AAZX01016347 DQ356057 ABC88772.1 KQ976464 KYM84415.1 NNAY01004759 OXU17455.1 NNAY01001310 OXU24399.1 KB031134 ELK02358.1 AXCN02002112 OAD52756.1 LBMM01002732 KMQ94422.1 AACZ04048256 DQ356062 ABC88777.1 AJFE02092719 AHZZ02020796 JTDY01004606 KOB67958.1 KQ981382 KYN42152.1 KE162362 EPQ07864.1 NBAG03000220 PNI78351.1 AK313628 BAG36388.1 M27717 M73720 M73716 M73717 M73718 M73719 AC092979 BC012613 S40234 DQ356061 ABC88776.1 CM001254 EHH16689.1 BABH01025123 BABH01025124 CM001277 EHH51597.1 JSUE03024456 AQIA01038222 AK088906 CH466530 BAC40646.1 EDL34896.1 J05118 CABD030024742 AQIB01076104 MK075163 AYV99566.1 KQ982959 KYQ48754.1 KB102464 ELK35370.1 KQ980658 KYN14778.1 GGMR01000219 MBY12838.1
KZ150003 PZC75272.1 KQ769940 OAD52755.1 GALX01004696 JAB63770.1 KZ288417 PBC26020.1 KK852482 KDR22872.1 GALX01004695 JAB63771.1 GALX01004555 JAB63911.1 KQ434938 KZC12084.1 AXCM01002791 DS232063 EDS33630.1 KQ414627 KOC67443.1 KQ976435 KYM87054.1 KQ977279 KYN04252.1 ADTU01026166 FX985401 BAX07414.1 GBYB01012027 JAG81794.1 ADTU01022957 NWSH01000796 PCG74167.1 KYN04096.1 KQ971348 EFA05749.1 APGK01045325 APGK01045326 KB741033 ENN74749.1 KB631976 ERL87560.1 GL888218 EGI64553.1 KQ435828 KOX71863.1 GFDF01005229 JAV08855.1 GFDF01005228 JAV08856.1 DQ356060 ABC88775.1 AXCM01005733 DQ356059 ABC88774.1 DQ356056 ABC88771.1 ABGA01105389 ABGA01105390 NDHI03003379 PNJ72804.1 KOC67445.1 PZC75275.1 AAZX01005059 AAZX01016347 DQ356057 ABC88772.1 KQ976464 KYM84415.1 NNAY01004759 OXU17455.1 NNAY01001310 OXU24399.1 KB031134 ELK02358.1 AXCN02002112 OAD52756.1 LBMM01002732 KMQ94422.1 AACZ04048256 DQ356062 ABC88777.1 AJFE02092719 AHZZ02020796 JTDY01004606 KOB67958.1 KQ981382 KYN42152.1 KE162362 EPQ07864.1 NBAG03000220 PNI78351.1 AK313628 BAG36388.1 M27717 M73720 M73716 M73717 M73718 M73719 AC092979 BC012613 S40234 DQ356061 ABC88776.1 CM001254 EHH16689.1 BABH01025123 BABH01025124 CM001277 EHH51597.1 JSUE03024456 AQIA01038222 AK088906 CH466530 BAC40646.1 EDL34896.1 J05118 CABD030024742 AQIB01076104 MK075163 AYV99566.1 KQ982959 KYQ48754.1 KB102464 ELK35370.1 KQ980658 KYN14778.1 GGMR01000219 MBY12838.1
Proteomes
UP000053240
UP000007151
UP000218220
UP000002358
UP000005203
UP000242457
+ More
UP000027135 UP000076502 UP000075883 UP000002320 UP000053825 UP000078540 UP000078542 UP000005205 UP000233220 UP000233040 UP000007266 UP000019118 UP000030742 UP000075885 UP000007755 UP000008225 UP000233020 UP000076408 UP000053105 UP000001595 UP000215335 UP000010552 UP000075886 UP000036403 UP000002277 UP000240080 UP000189706 UP000028761 UP000037510 UP000078541 UP000233180 UP000005640 UP000233120 UP000005204 UP000009130 UP000006718 UP000233100 UP000000589 UP000001519 UP000029965 UP000233080 UP000075809 UP000078492 UP000252040 UP000233200
UP000027135 UP000076502 UP000075883 UP000002320 UP000053825 UP000078540 UP000078542 UP000005205 UP000233220 UP000233040 UP000007266 UP000019118 UP000030742 UP000075885 UP000007755 UP000008225 UP000233020 UP000076408 UP000053105 UP000001595 UP000215335 UP000010552 UP000075886 UP000036403 UP000002277 UP000240080 UP000189706 UP000028761 UP000037510 UP000078541 UP000233180 UP000005640 UP000233120 UP000005204 UP000009130 UP000006718 UP000233100 UP000000589 UP000001519 UP000029965 UP000233080 UP000075809 UP000078492 UP000252040 UP000233200
Interpro
Gene 3D
ProteinModelPortal
A0A194RGY2
A0A212F6I3
A0A2A4J3C4
A0A2W1BJL8
K7IN13
A0A310SEP0
+ More
A0A088A4S2 V5G180 A0A2A3E2U1 A0A067RGG5 V5I8H1 V5I8J9 A0A154PKT4 A0A182LX23 B0WS12 A0A0L7R9B9 A0A195BNW8 A0A195CUQ5 A0A158NTV3 A0A2K6V6C3 A0A2K5PME4 A0A1V1FIW8 A0A0C9RFV3 A0A158NQ44 A0A2A4JQ49 A0A195CTS9 D2A3Y2 N6T3L7 U4UC85 K7IQY4 A0A182P9F1 F4WMS6 F7HXH8 A0A2K5EUS2 A0A182YDB7 A0A0M8ZXY7 A0A1L8DR93 A0A1L8DQV7 A1XGA1 A0A182LT40 A1XGA0 A1XG97 H2PBP8 A0A0L7R992 A0A2W1BNU0 K7JA32 A1XG98 A0A195BHV6 A0A232EGG7 A0A232F1J1 K7IY61 L5JVM8 A0A182QAV5 A0A310SCR6 A0A0J7KVA1 A0A2I3TU16 A1XGA3 A0A2R9CAY4 A0A3Q0D5I3 A0A096P028 A0A0L7KXG1 H2QNJ6 A0A195FPB2 S7MU95 A0A2K6KUE9 A0A2R9CLS4 A0A2J8P2V4 B2R941 P15088 A0A158NQ85 A1XGA2 A0A2K6C4D9 G7MJJ7 H9J5L1 G7NZN8 F7GFE7 A0A2K5WCY7 Q542E3 P15089 G3S533 A0A2K6LW11 A0A2K6LVY0 A0A0D9RG95 A0A2K5JGG1 A0A3G5BIF5 A0A2K5CNU8 A0A151WLL6 L5MAR4 A0A151J021 A0A2S2N6Y1 A0A2K5R360 A0A2K5CNV4 A0A341B9Q3 A0A2K6NNH6
A0A088A4S2 V5G180 A0A2A3E2U1 A0A067RGG5 V5I8H1 V5I8J9 A0A154PKT4 A0A182LX23 B0WS12 A0A0L7R9B9 A0A195BNW8 A0A195CUQ5 A0A158NTV3 A0A2K6V6C3 A0A2K5PME4 A0A1V1FIW8 A0A0C9RFV3 A0A158NQ44 A0A2A4JQ49 A0A195CTS9 D2A3Y2 N6T3L7 U4UC85 K7IQY4 A0A182P9F1 F4WMS6 F7HXH8 A0A2K5EUS2 A0A182YDB7 A0A0M8ZXY7 A0A1L8DR93 A0A1L8DQV7 A1XGA1 A0A182LT40 A1XGA0 A1XG97 H2PBP8 A0A0L7R992 A0A2W1BNU0 K7JA32 A1XG98 A0A195BHV6 A0A232EGG7 A0A232F1J1 K7IY61 L5JVM8 A0A182QAV5 A0A310SCR6 A0A0J7KVA1 A0A2I3TU16 A1XGA3 A0A2R9CAY4 A0A3Q0D5I3 A0A096P028 A0A0L7KXG1 H2QNJ6 A0A195FPB2 S7MU95 A0A2K6KUE9 A0A2R9CLS4 A0A2J8P2V4 B2R941 P15088 A0A158NQ85 A1XGA2 A0A2K6C4D9 G7MJJ7 H9J5L1 G7NZN8 F7GFE7 A0A2K5WCY7 Q542E3 P15089 G3S533 A0A2K6LW11 A0A2K6LVY0 A0A0D9RG95 A0A2K5JGG1 A0A3G5BIF5 A0A2K5CNU8 A0A151WLL6 L5MAR4 A0A151J021 A0A2S2N6Y1 A0A2K5R360 A0A2K5CNV4 A0A341B9Q3 A0A2K6NNH6
PDB
5MRV
E-value=4.26837e-20,
Score=236
Ontologies
GO
Topology
Subcellular location
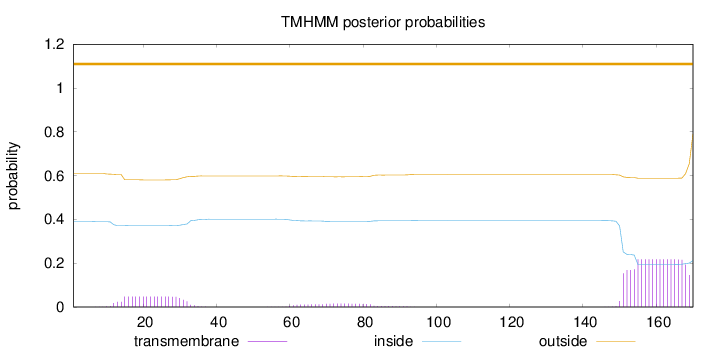
Length:
170
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
5.1002
Exp number, first 60 AAs:
0.91849
Total prob of N-in:
0.38997
outside
1 - 170
Population Genetic Test Statistics
Pi
172.861931
Theta
162.908028
Tajima's D
-1.330247
CLR
3.953233
CSRT
0.0825958702064897
Interpretation
Uncertain
