Gene
KWMTBOMO15018 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA004797
Annotation
PREDICTED:_zinc_carboxypeptidase-like_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 3.007
Sequence
CDS
ATGAATTACAAAATTTATTATTTTCTTCTAATAACTTTTAGCGTAGTTAACGCTGTTAAAGATTACAATGGATACAAAGTATACAGAGTGGTACCGAGGACTGAAGATGATGTGAAGCTTTTAGAAGAATTCAGGAGGACGTCGGTGGGAGAGTTCTGGGATGATAATCTGGTTGTTAATTATAGAGTAAACATGATGGTGCCAGGTGATATGCAGGGGTTATTCCTGAATTCCACTCGTCAAACTATGATTGAGGTTGAAGAAGTGATACCGGATTTACAAAAGGCAATAGAGGACCAATTAAAACCCGCAACCGGATTTAGAAGTTCGGAAACCTTTCATGGCTATAGCTGGGACAGGTATCACAATTTAGAAACAATAAATACTTGGATGAATACCTTAGTACAAAATTATCCTGGTATAGTACGACCAGTTGTTATGGGAAGATCAGTTGAGAACAGAGAAATTAGAGGTATCATCATCAATTATAAGCCAGAAAAGACTAATAAGCTAATGGGGATCCTGGAAGGAACTCTTCATGCTAGAGAATGGATCGCACCGGCTACTGTAACATGGATAGTCAAAGAGTTTTTAACTAGTACAGACCCGCAAGTCAGGGCAATGGCTGAGAACATTGAATGGCATGTATTTCCAGTGGTTAATCCTGATGGATATCACTATAGCTTCACAACTCATCGGATGTGGAGGAAGAATCGTAGCCCTGAAAGTTTTGGAACCTGTGAGGGAGCCGATGACGATATGAGCCATGGAGTAGATTTGAATAGAAACTTTGATTTTGTTTGGATGTCTGTAGGAGCATCAAATAATCCTTGTTCTAACACTTTTGCGGGACCGAGGGCATTTTCTGAACCCGAATCTAGAGCCATATCTCAGTATGTCCTCAACTTGAAAGAACAGGGAGAGATAATATACTATCTTGCTTACCACTCCTATACTCAACTCATAGTGATTCCGTACAGTCATGTGACTGGATCTGACGTTCTGCTTGCCAATAATTATGCTGATATGTATGAAATTGGTATACGAAGTGCCGATGCTATCGCAAAAAGATACGGTACAGTTTATAGGGTCGGCGTTTCGTCGGAAATCCTGTATCCCATTAGTGGCGGCAGTTTTGATTGGGTCAAGAAAGTGGCAGATGTACCTATCACTTTCTTGATTGAGCTGCGTGATCTTGGAGAATACGGGTTTCTTCTTCCGCCAGAACAGATCATCCCCAACAACCTTGAAACTATGGATGGACTTATCGAGATGGATCGCGTGACTAAGCTTCTTGGCTATTACTCGGCTGGTGCAATTAGTAAAGTAATTTCTTTGTCTGTTTTAATTTTGTGCTCAATGGTTATAGTTTTATTTAATTAA
Protein
MNYKIYYFLLITFSVVNAVKDYNGYKVYRVVPRTEDDVKLLEEFRRTSVGEFWDDNLVVNYRVNMMVPGDMQGLFLNSTRQTMIEVEEVIPDLQKAIEDQLKPATGFRSSETFHGYSWDRYHNLETINTWMNTLVQNYPGIVRPVVMGRSVENREIRGIIINYKPEKTNKLMGILEGTLHAREWIAPATVTWIVKEFLTSTDPQVRAMAENIEWHVFPVVNPDGYHYSFTTHRMWRKNRSPESFGTCEGADDDMSHGVDLNRNFDFVWMSVGASNNPCSNTFAGPRAFSEPESRAISQYVLNLKEQGEIIYYLAYHSYTQLIVIPYSHVTGSDVLLANNYADMYEIGIRSADAIAKRYGTVYRVGVSSEILYPISGGSFDWVKKVADVPITFLIELRDLGEYGFLLPPEQIIPNNLETMDGLIEMDRVTKLLGYYSAGAISKVISLSVLILCSMVIVLFN
Summary
Uniprot
A0A2A4J213
A0A2W1BR80
A0A194RM68
A0A2H1VG25
A0A194PFK4
A0A194RHN6
+ More
A0A212EX84 H9BVM4 A0A1E1VZW2 A0A212F6G8 A0A2A4J379 A0A2H1WTP5 A0A2W1BLV7 H9J5K8 B0WS12 A0A182LT40 A0A182QAV5 A0A182YDB7 A0A182QY48 A0A182JEM2 W5JK64 V9QNN0 B0WS11 A0A3G5BIF5 V9QMH2 A0A182RFC6 A0A1S4F4N5 Q7Q316 A0A182H3R1 A0A182TPH8 A0A1V1FIW8 Q6J6D5 A0A182UYU3 A0A310SCR6 Q5TPZ3 A0A182V379 A0A2C9GR29 A0A1I8JW63 A0A182LPK9 K7IN13 A0A182TX13 A0A182MXY6 A0A182WF36 T1DQS4 A0A154PJT5 A0A0M8ZXY7 A0A0C9RFV3 A0A182FUU7 A0A336MJB3 A0A2A4JQ49 F4WJ50 K7IQY4 W5JN76 A0A2P8YM65 A0A182K4R6 Q0QWG1 A0A154PKT4 A0A336MCF7 A0A182P9F0 K7IY61 A0A1I8JW39 W5J673 A0A067RG70 A0A182LPL5 A0A2C9GQU4 A0A2C9GS83 A7UUL1 A0A232F435 A0A182L0M0 A0A0L7R9B9 Q16YB7 A0A1S4FJX8 Q6J6D8 A0A2A3E2U1 A0A2H1X3V0 A0A195FPR9 V5I8H1 D2A3Y2 A0A151WLL6 A0A1W4WZ68 A0A182QXZ6 A0A195CTS9 A0A084VIH2 A0A310SEP0 A0A182UHK5 A0A2J7REL0 A0A232F1J1 V5G180 A0A1W4XFF7 A0A195BNW8 A0A232EGG7 Q5QBL4 A0A2J7REK5 A0A3F2Z144 A1XG98 A0A0L7R992 A1XGA1 A0A182GL89
A0A212EX84 H9BVM4 A0A1E1VZW2 A0A212F6G8 A0A2A4J379 A0A2H1WTP5 A0A2W1BLV7 H9J5K8 B0WS12 A0A182LT40 A0A182QAV5 A0A182YDB7 A0A182QY48 A0A182JEM2 W5JK64 V9QNN0 B0WS11 A0A3G5BIF5 V9QMH2 A0A182RFC6 A0A1S4F4N5 Q7Q316 A0A182H3R1 A0A182TPH8 A0A1V1FIW8 Q6J6D5 A0A182UYU3 A0A310SCR6 Q5TPZ3 A0A182V379 A0A2C9GR29 A0A1I8JW63 A0A182LPK9 K7IN13 A0A182TX13 A0A182MXY6 A0A182WF36 T1DQS4 A0A154PJT5 A0A0M8ZXY7 A0A0C9RFV3 A0A182FUU7 A0A336MJB3 A0A2A4JQ49 F4WJ50 K7IQY4 W5JN76 A0A2P8YM65 A0A182K4R6 Q0QWG1 A0A154PKT4 A0A336MCF7 A0A182P9F0 K7IY61 A0A1I8JW39 W5J673 A0A067RG70 A0A182LPL5 A0A2C9GQU4 A0A2C9GS83 A7UUL1 A0A232F435 A0A182L0M0 A0A0L7R9B9 Q16YB7 A0A1S4FJX8 Q6J6D8 A0A2A3E2U1 A0A2H1X3V0 A0A195FPR9 V5I8H1 D2A3Y2 A0A151WLL6 A0A1W4WZ68 A0A182QXZ6 A0A195CTS9 A0A084VIH2 A0A310SEP0 A0A182UHK5 A0A2J7REL0 A0A232F1J1 V5G180 A0A1W4XFF7 A0A195BNW8 A0A232EGG7 Q5QBL4 A0A2J7REK5 A0A3F2Z144 A1XG98 A0A0L7R992 A1XGA1 A0A182GL89
Pubmed
EMBL
NWSH01003708
PCG65899.1
KZ150003
PZC75270.1
KQ460205
KPJ17076.1
+ More
ODYU01002374 SOQ39789.1 KQ459604 KPI92156.1 KPJ17077.1 AGBW02011823 OWR46093.1 JQ081295 AFD99126.1 GDQN01010791 JAT80263.1 AGBW02010033 OWR49318.1 PCG65900.1 ODYU01010985 SOQ56440.1 PZC75271.1 BABH01025136 BABH01025137 DS232063 EDS33630.1 AXCM01005733 AXCN02002112 ADMH02000883 ETN64767.1 ATLV01013356 KF707629 KE524854 AHC32031.1 KFB37767.1 EDS33629.1 MK075163 AYV99566.1 KF707628 AHC32030.1 AAAB01008966 EAA13019.4 JXUM01024706 KQ560699 KXJ81228.1 FX985401 BAX07414.1 AY590492 AAT36730.1 KQ769940 OAD52756.1 EAL39491.3 APCN01001947 GAMD01002289 JAA99301.1 KQ434938 KZC12083.1 KQ435828 KOX71863.1 GBYB01012027 JAG81794.1 UFQT01001101 SSX28923.1 NWSH01000796 PCG74167.1 GL888181 EGI65820.1 ETN64768.1 PYGN01000496 PSN45348.1 DQ196074 ABA29654.1 KZC12084.1 UFQT01000916 SSX27975.1 ADMH02002111 ETN58893.1 KK852482 KDR22871.1 APCN01000778 AAAB01008964 EDO63564.1 NNAY01001025 OXU25435.1 KQ414627 KOC67443.1 CH477519 EAT39609.1 AY590489 AAT36727.1 KZ288417 PBC26020.1 ODYU01013306 SOQ60013.1 KQ981382 KYN42302.1 GALX01004695 JAB63771.1 KQ971348 EFA05749.1 KQ982959 KYQ48754.1 AXCN02000807 KQ977279 KYN04096.1 KFB37766.1 OAD52755.1 NEVH01004959 PNF39269.1 NNAY01001310 OXU24399.1 GALX01004696 JAB63770.1 KQ976435 KYM87054.1 NNAY01004759 OXU17455.1 AY752797 UFQS01000369 UFQT01000369 AAV84210.1 SSX03243.1 SSX23609.1 PNF39268.1 DQ356057 ABC88772.1 KOC67445.1 DQ356060 ABC88775.1 JXUM01071336 KQ562658 KXJ75389.1
ODYU01002374 SOQ39789.1 KQ459604 KPI92156.1 KPJ17077.1 AGBW02011823 OWR46093.1 JQ081295 AFD99126.1 GDQN01010791 JAT80263.1 AGBW02010033 OWR49318.1 PCG65900.1 ODYU01010985 SOQ56440.1 PZC75271.1 BABH01025136 BABH01025137 DS232063 EDS33630.1 AXCM01005733 AXCN02002112 ADMH02000883 ETN64767.1 ATLV01013356 KF707629 KE524854 AHC32031.1 KFB37767.1 EDS33629.1 MK075163 AYV99566.1 KF707628 AHC32030.1 AAAB01008966 EAA13019.4 JXUM01024706 KQ560699 KXJ81228.1 FX985401 BAX07414.1 AY590492 AAT36730.1 KQ769940 OAD52756.1 EAL39491.3 APCN01001947 GAMD01002289 JAA99301.1 KQ434938 KZC12083.1 KQ435828 KOX71863.1 GBYB01012027 JAG81794.1 UFQT01001101 SSX28923.1 NWSH01000796 PCG74167.1 GL888181 EGI65820.1 ETN64768.1 PYGN01000496 PSN45348.1 DQ196074 ABA29654.1 KZC12084.1 UFQT01000916 SSX27975.1 ADMH02002111 ETN58893.1 KK852482 KDR22871.1 APCN01000778 AAAB01008964 EDO63564.1 NNAY01001025 OXU25435.1 KQ414627 KOC67443.1 CH477519 EAT39609.1 AY590489 AAT36727.1 KZ288417 PBC26020.1 ODYU01013306 SOQ60013.1 KQ981382 KYN42302.1 GALX01004695 JAB63771.1 KQ971348 EFA05749.1 KQ982959 KYQ48754.1 AXCN02000807 KQ977279 KYN04096.1 KFB37766.1 OAD52755.1 NEVH01004959 PNF39269.1 NNAY01001310 OXU24399.1 GALX01004696 JAB63770.1 KQ976435 KYM87054.1 NNAY01004759 OXU17455.1 AY752797 UFQS01000369 UFQT01000369 AAV84210.1 SSX03243.1 SSX23609.1 PNF39268.1 DQ356057 ABC88772.1 KOC67445.1 DQ356060 ABC88775.1 JXUM01071336 KQ562658 KXJ75389.1
Proteomes
UP000218220
UP000053240
UP000053268
UP000007151
UP000005204
UP000002320
+ More
UP000075883 UP000075886 UP000076408 UP000075880 UP000000673 UP000030765 UP000075900 UP000007062 UP000069940 UP000249989 UP000075902 UP000075903 UP000075840 UP000076407 UP000075882 UP000002358 UP000075884 UP000075920 UP000076502 UP000053105 UP000069272 UP000007755 UP000245037 UP000075881 UP000075885 UP000027135 UP000215335 UP000053825 UP000008820 UP000242457 UP000078541 UP000007266 UP000075809 UP000192223 UP000078542 UP000235965 UP000078540
UP000075883 UP000075886 UP000076408 UP000075880 UP000000673 UP000030765 UP000075900 UP000007062 UP000069940 UP000249989 UP000075902 UP000075903 UP000075840 UP000076407 UP000075882 UP000002358 UP000075884 UP000075920 UP000076502 UP000053105 UP000069272 UP000007755 UP000245037 UP000075881 UP000075885 UP000027135 UP000215335 UP000053825 UP000008820 UP000242457 UP000078541 UP000007266 UP000075809 UP000192223 UP000078542 UP000235965 UP000078540
Interpro
Gene 3D
ProteinModelPortal
A0A2A4J213
A0A2W1BR80
A0A194RM68
A0A2H1VG25
A0A194PFK4
A0A194RHN6
+ More
A0A212EX84 H9BVM4 A0A1E1VZW2 A0A212F6G8 A0A2A4J379 A0A2H1WTP5 A0A2W1BLV7 H9J5K8 B0WS12 A0A182LT40 A0A182QAV5 A0A182YDB7 A0A182QY48 A0A182JEM2 W5JK64 V9QNN0 B0WS11 A0A3G5BIF5 V9QMH2 A0A182RFC6 A0A1S4F4N5 Q7Q316 A0A182H3R1 A0A182TPH8 A0A1V1FIW8 Q6J6D5 A0A182UYU3 A0A310SCR6 Q5TPZ3 A0A182V379 A0A2C9GR29 A0A1I8JW63 A0A182LPK9 K7IN13 A0A182TX13 A0A182MXY6 A0A182WF36 T1DQS4 A0A154PJT5 A0A0M8ZXY7 A0A0C9RFV3 A0A182FUU7 A0A336MJB3 A0A2A4JQ49 F4WJ50 K7IQY4 W5JN76 A0A2P8YM65 A0A182K4R6 Q0QWG1 A0A154PKT4 A0A336MCF7 A0A182P9F0 K7IY61 A0A1I8JW39 W5J673 A0A067RG70 A0A182LPL5 A0A2C9GQU4 A0A2C9GS83 A7UUL1 A0A232F435 A0A182L0M0 A0A0L7R9B9 Q16YB7 A0A1S4FJX8 Q6J6D8 A0A2A3E2U1 A0A2H1X3V0 A0A195FPR9 V5I8H1 D2A3Y2 A0A151WLL6 A0A1W4WZ68 A0A182QXZ6 A0A195CTS9 A0A084VIH2 A0A310SEP0 A0A182UHK5 A0A2J7REL0 A0A232F1J1 V5G180 A0A1W4XFF7 A0A195BNW8 A0A232EGG7 Q5QBL4 A0A2J7REK5 A0A3F2Z144 A1XG98 A0A0L7R992 A1XGA1 A0A182GL89
A0A212EX84 H9BVM4 A0A1E1VZW2 A0A212F6G8 A0A2A4J379 A0A2H1WTP5 A0A2W1BLV7 H9J5K8 B0WS12 A0A182LT40 A0A182QAV5 A0A182YDB7 A0A182QY48 A0A182JEM2 W5JK64 V9QNN0 B0WS11 A0A3G5BIF5 V9QMH2 A0A182RFC6 A0A1S4F4N5 Q7Q316 A0A182H3R1 A0A182TPH8 A0A1V1FIW8 Q6J6D5 A0A182UYU3 A0A310SCR6 Q5TPZ3 A0A182V379 A0A2C9GR29 A0A1I8JW63 A0A182LPK9 K7IN13 A0A182TX13 A0A182MXY6 A0A182WF36 T1DQS4 A0A154PJT5 A0A0M8ZXY7 A0A0C9RFV3 A0A182FUU7 A0A336MJB3 A0A2A4JQ49 F4WJ50 K7IQY4 W5JN76 A0A2P8YM65 A0A182K4R6 Q0QWG1 A0A154PKT4 A0A336MCF7 A0A182P9F0 K7IY61 A0A1I8JW39 W5J673 A0A067RG70 A0A182LPL5 A0A2C9GQU4 A0A2C9GS83 A7UUL1 A0A232F435 A0A182L0M0 A0A0L7R9B9 Q16YB7 A0A1S4FJX8 Q6J6D8 A0A2A3E2U1 A0A2H1X3V0 A0A195FPR9 V5I8H1 D2A3Y2 A0A151WLL6 A0A1W4WZ68 A0A182QXZ6 A0A195CTS9 A0A084VIH2 A0A310SEP0 A0A182UHK5 A0A2J7REL0 A0A232F1J1 V5G180 A0A1W4XFF7 A0A195BNW8 A0A232EGG7 Q5QBL4 A0A2J7REK5 A0A3F2Z144 A1XG98 A0A0L7R992 A1XGA1 A0A182GL89
PDB
5MRV
E-value=9.78171e-61,
Score=592
Ontologies
GO
Topology
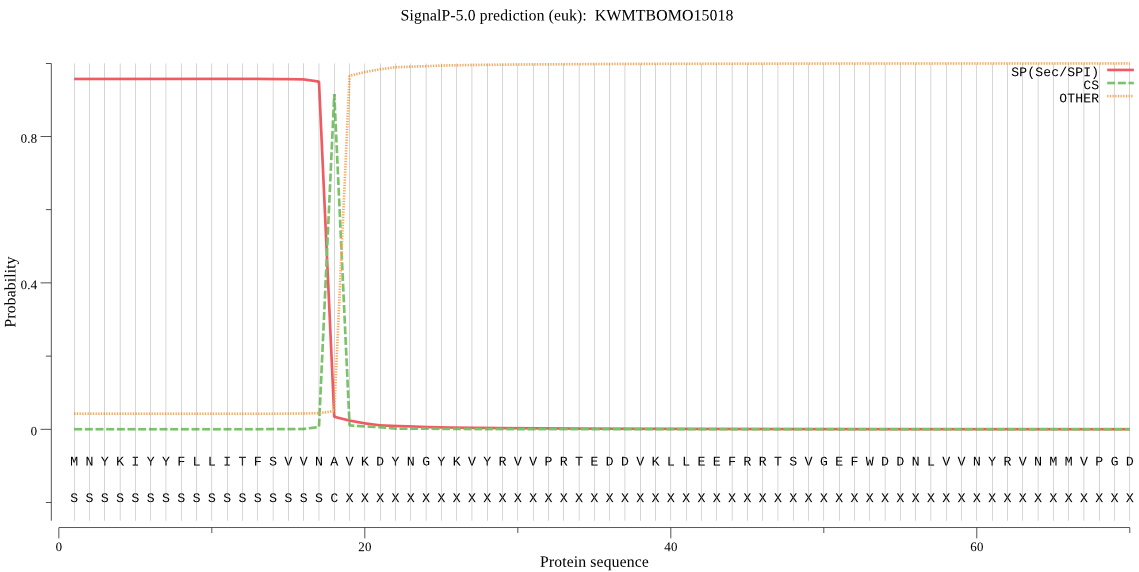
SignalP
Position: 1 - 18,
Likelihood: 0.956876
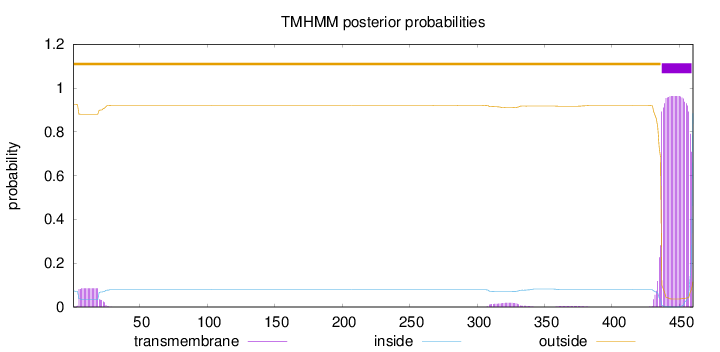
Length:
460
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
24.11333
Exp number, first 60 AAs:
1.44482
Total prob of N-in:
0.07434
outside
1 - 436
TMhelix
437 - 459
inside
460 - 460
Population Genetic Test Statistics
Pi
160.120278
Theta
179.979538
Tajima's D
-0.741883
CLR
202.805913
CSRT
0.187390630468477
Interpretation
Uncertain
