Pre Gene Modal
BGIBMGA004872
Annotation
PREDICTED:_eukaryotic_translation_initiation_factor_2-alpha_kinase_4_[Amyelois_transitella]
Full name
eIF-2-alpha kinase GCN2
Alternative Name
Eukaryotic translation initiation factor 2-alpha kinase 4
GCN2-like protein
GCN2-like protein
Location in the cell
Nuclear Reliability : 2.939
Sequence
CDS
ATGGCTGAAGAGAGCTACGAGGAGAGACAAAATAATGAGCTAACCGCCTTACAGGCTATATATGGTGATGCTGTTATTGATAATCGCGAAAACATTGCTTGGAAAGTATGGCGTCCGCTCGATATAATGTTGACGTTGAATCCACTTCACAACTCAGATATACAAGGTGTTCACTGCTCTGTAACATTGCATTTAAAATGTTGCAATAATTATCCTGACAAGCCACCAAGTCTAGCCATACACAAGATGCATGGATTATCCACAGACAATGCCACCAAATTATTGGCTGAATTGGAGACATTGTCACATCAATTGTGTGGTGAAGTTATGATATTCCAACTAGCCCAGCATGCACAGCAATTTCTGCACGATCACAATAAGCCTATATTGTCGTTTTACGATGAAATGCTGAAGCAGAAGAATGAAATGGAGAAACTGAAACTTCATGATTTACAAACCAAAGAGGAGGAAGAACGCCAAAAAATCAAAGATGAGATACAACGTCGTCAGGAGATACTCAAAGGTGCCGGAAGACAGCGTGTCCGAGTCTCCAGCCTCTGCCATGATAACAGCACGGAACAGGACTGCACTGATGAAAAGGTGGATGCACCAGAGCTTGTATTCTATGCGGACGATAAAATGTCGCCAGTAAAGAAATCAACACGTCTTCGAAACATGGAGGTTCCGTGCACGTGTAACACGCGAGGAGTGCAAGTGCTCAGAGTGACGCAACGGAACAACAAGAAGGTTTACATAGGGAGCTGTTTAGGACATTCATCGACCGGCGCGACAACATATCTAGCGATAGACGACGACACAGGGGAAAGTCTTATCGCAAAGAAGTGGAGCATCCCACCGGCTTCAGACTTCCAAACGAGAAACAAACAGCTTACATCGATACAGCAAGACGTGAAGTCGCTTGCCCGACTCAGCAACGCCTCCTTGATCCCATACGTTGCAATGGAGATGTGTAAAGAAGTAAACAAACGAACGTCAAAACAGTGCATTTATATATTCAGAAGTTTCGTTCTAGGGACGTCCTTGAAGAATCTAGTGAAAAAGTTGAGTGGTTTCAGTGATTGCTTTGAGGTTTTGGGCCTCTTGAGACATGTGGCTGACGGTGTTTTGAGTGCGTTAAACGAATTGCATGCAGCGAATATAATACATAGGGATGTTAGAAGTGAAAATGTATTCGTCGATGACGTTGGCCAAGTGAAGTTGGTTGGTGCTAGCTTGGACACCAGGTTGGCTGAAATGTTGGACAATGATAACTATTGTGATAAGCAAACCCCAGCCCAAGATATCTATGCGACCGGACAACTTCTGTTATCCATTGTTTGCAAAGAAAAATCGTATACAGAAATACCGCAGGAGCTCCCTTGCGTGGCAAAAGACTTCATATCCAGATGTTTAACAGAAGACGAACATTCGCAATGGTCGGCGAAGCAGTTGATGAGTCATTACTTCTTAGTCGAAGCTCCGACAAAGAAACCTTACGTCAAACGTGCCGACGACGGTGGAAGCGGTTCAGAGGACGACGACGCCGTCAAAAAGATTCGACACATGAGTTTTGCCAACAACGGGCTGTCGCGGCTGGACGCTGAGTTTGAGGTGCTAGCGTGGCTCGGGAAAGGCGCATTCGGTGATGTTTTGAAGGTAAAGAACAAACTGGACGGAGGTTTCTACGCGATAAAAAGAGTCCAATTGAATCCTGAAAGCGTTCAGCTCAACAAGAAGATAACAAGGGAAGTGAAACTCCTCTCCAGGCTCAACCACGAGAACGTAGTCCGCTATTACAACGCGTGGATCGAGTCGACAACTGAGCCGATAGACTGTGAGGAATCTCCCACCAAAACTCCTACCCGGAAACCCAGCTTGGAAGGTATTGTCGCGAAACTGGGACAAGAGGTCAAAGTGGAGTGGTCCATGTCAGAGGGACCGGCGCAGATGACGAATTCCACATCGGACACCGAAGATAGCGATGACGATGAACCGGATCCGTGGTTTAATATCATCAGCCCCGGTGACGAATCGAGCGAACTTATCGAATTCGAAAAGACCTCTGACGACGAAGAATCTCAGACGTTACCATCGGAAGACGTTGTCGACGCGCCGAGACCTCAATCCGAGCGTCTCAAGCAAGTCCTATACATCCAAATGGAATTTTGCGAGAAGCATACATTGAGACAAGCCATAGACAACGGCTTGTTCCAGGAACATTTCAGGGCGTGGAGGCTGTTCCGGGAGATCGTCGAGGGTCTGGCCCACGTGCATCAGAGAGGCATGATACACAGGGACCTCAAGCCTGTGAATATATTCTTGGACTCGAACGACCATGTCAAGATTGGAGACTTCGGACTAGCAACTAAGGCGTTCACGGGACTGCCGGTTGAAGATAAAACGAAACAAGAAGAAATAGGCGGTTCGCTAACTGGGCAAATAGGAACAGCCCTATACGTAGCTCCCGAACTACTCCAGTCTGCTACAAAGGTCATATACAATCAAAAAGTGGACATATATTCATTAGGCATAATATTGTTTGAAATGTTCCATCCGCCCCTCGAAACTGGAATGGAGAGAATGATGGTGCTCACCAATTTGAGGAGCAAGGATATCGTCATACCGAATAAGTTTGATGTCGACGAAAATGTGAAGCAGATTCATGTCATAAGATGGTTGCTGAACCACGAGGCGAGCCTGCGGCCCACGTGCGCGGAGCTGCTGGCCAGCGAGCACGTGCCGCGCGCCGTGCCCGAGGGCGCGCTGGCCGGGCTGCTGTCGCACGCGCTCAGCGACCGCACGGCGCGCGGCTACCAGCGCCTCGTGGCCGCCTGCCTCGACCAGAGGCCGTCCGCAGCCGAGGACTACACCTACCACAACGGGATGAAGACCAAGCCCGCCCGTCTCCTGGAGTTCGTCAAGGATGCTGTTGTCAAGGTATTCAAAAGTCACGGAGCTTTAGAGTTCGCTCCGCCGCTCCTTATACCGAGAGCGAGAGCCTGGGACCAGTATCCTAACGCTGTCAAAGTCATGACGTCATCGGGCTCGGTCTGTCATTTACCGCACGACTTGAGGCTGCCTTTCGCCAGGCACATAGCATACTCCGGTATAAAAAATCTCCGTCGTTTTATAGTCGATAGGGTTTATAGAGAGAAGCACGTTCAAGGTTTTCACCCGAAGGAAATTCTTGAATGTGCCTACGATATCGTCACCCCTAAAACTGACTCTCTATGGCCGGACGCAGAATTAATAGAGGTCGCAAGTAGAGCGGCATCGGAGTGTTCCCTCAAAGTGTCGATACAACTAAATCACACGGAACTGATTCGCGTCCTGCTACAGTCGTGCGGGGTACCGACAGACAAACAGGCTGATATATACCCTGTTTTGGTCGACGTCAGCTTCGGTCGCATAACGAGTCTGCAACTTCAAACACACTTGACGTCGCTCTGTATAACGAATCGTGACGTCACGAACCTGGTCCGCCTGATGGAGGCGGATGCGCCGGTCCAAGAGCTCCGGGAGCTGGTCACGCCTCTCGTCAAACAAGCGAAGTGCAGTAGGATCGCGAGTCAGGCCGTGCACGACCTGGAAGCCGTGTACAGGAACGCCAAGGCGCTGGGCTGTGAGTGCCCGATGACGGTGGCTCCGCTCTCGGCGTACAACGTGACGCAGCACAGCGGCGTGTTCTGGCAGATGTCCGTCGTGCGAAGCGAACAGAAGTCGATCACCAAACGACGTTCCGGGGACCTTATTGCGGCTGGCGGAAGATACGACGCTCTGGTCAAGGAGTTCTGGAATGTAGCGAGCGCGGAAAACGATCAAGACAGCGCCGATTTGAATGTGTGTTCGGTCGGGTTCTCGATGTCCCTCGAACGGATGGCGGCCTTCCTCAACAAACTGGAGACGCAAACGAACGTGCCGATCAAGAGTAACGAGACGCCGCTGGTGCACGTGCGCGTGTGGGGCGCGGTGGGCAGCGGGCGGGAGGGCGCGGCGGCGGCGCGGCGGGCGCAGCTGGCGCGCGATCTGTGGGCGGGCGGGCTGCGAGCCGGGCTGCACGCGGGGGACGCGCCCGACCTCGCGCCCGGCGCCGTGCTGGCCGAGCACGACCCGCCCCTCGTCCGGCTGTCCTGCTGGGACCAGGACAGGATCCGGGAACACAGGGTGCCCTACGTGGATGTTGTCGATTTCATCAAACAAAAAATTATCGGCGACTCGTCTCGGAATCCTGAAGGTTCTAGCACGAACCGATCGATAAGCTGGTCCGAAAGCTCCGAAAAATACAACAGTCCGAATATATCGGTCACTTTCATAACGGCCAGCGACAGAATGACGAAAAACTCGAAGAGGCACTGCGAGAATCAGATAAACACTCAAGTGACGTCGATTCTGGTAAGTCTGGGCCTCCTGCCGGTCATGGCGAGGGTGCGCGTCAACGTACTAGCCGTGGCCTGCGACTCCGCCTGCGTCAGGTACATGGCCTCGCAGCTGACGGCGCCATTAGATGCCGATGATCTGTCGCAAGCCTTCAGAAACGTTACGGAAATGTTCCCCAAACGTCAGGGCCTAATAGACGAAGCTCGCTCCGAGCTTTCGAGCTTATTGAAACAAACCAGCCCGAGTCGGAACAACGAAGAGACTCAAATATACGCCCTATATTCAATACCGGATTCATTGTGTCGACTTATTACTTGA
Protein
MAEESYEERQNNELTALQAIYGDAVIDNRENIAWKVWRPLDIMLTLNPLHNSDIQGVHCSVTLHLKCCNNYPDKPPSLAIHKMHGLSTDNATKLLAELETLSHQLCGEVMIFQLAQHAQQFLHDHNKPILSFYDEMLKQKNEMEKLKLHDLQTKEEEERQKIKDEIQRRQEILKGAGRQRVRVSSLCHDNSTEQDCTDEKVDAPELVFYADDKMSPVKKSTRLRNMEVPCTCNTRGVQVLRVTQRNNKKVYIGSCLGHSSTGATTYLAIDDDTGESLIAKKWSIPPASDFQTRNKQLTSIQQDVKSLARLSNASLIPYVAMEMCKEVNKRTSKQCIYIFRSFVLGTSLKNLVKKLSGFSDCFEVLGLLRHVADGVLSALNELHAANIIHRDVRSENVFVDDVGQVKLVGASLDTRLAEMLDNDNYCDKQTPAQDIYATGQLLLSIVCKEKSYTEIPQELPCVAKDFISRCLTEDEHSQWSAKQLMSHYFLVEAPTKKPYVKRADDGGSGSEDDDAVKKIRHMSFANNGLSRLDAEFEVLAWLGKGAFGDVLKVKNKLDGGFYAIKRVQLNPESVQLNKKITREVKLLSRLNHENVVRYYNAWIESTTEPIDCEESPTKTPTRKPSLEGIVAKLGQEVKVEWSMSEGPAQMTNSTSDTEDSDDDEPDPWFNIISPGDESSELIEFEKTSDDEESQTLPSEDVVDAPRPQSERLKQVLYIQMEFCEKHTLRQAIDNGLFQEHFRAWRLFREIVEGLAHVHQRGMIHRDLKPVNIFLDSNDHVKIGDFGLATKAFTGLPVEDKTKQEEIGGSLTGQIGTALYVAPELLQSATKVIYNQKVDIYSLGIILFEMFHPPLETGMERMMVLTNLRSKDIVIPNKFDVDENVKQIHVIRWLLNHEASLRPTCAELLASEHVPRAVPEGALAGLLSHALSDRTARGYQRLVAACLDQRPSAAEDYTYHNGMKTKPARLLEFVKDAVVKVFKSHGALEFAPPLLIPRARAWDQYPNAVKVMTSSGSVCHLPHDLRLPFARHIAYSGIKNLRRFIVDRVYREKHVQGFHPKEILECAYDIVTPKTDSLWPDAELIEVASRAASECSLKVSIQLNHTELIRVLLQSCGVPTDKQADIYPVLVDVSFGRITSLQLQTHLTSLCITNRDVTNLVRLMEADAPVQELRELVTPLVKQAKCSRIASQAVHDLEAVYRNAKALGCECPMTVAPLSAYNVTQHSGVFWQMSVVRSEQKSITKRRSGDLIAAGGRYDALVKEFWNVASAENDQDSADLNVCSVGFSMSLERMAAFLNKLETQTNVPIKSNETPLVHVRVWGAVGSGREGAAAARRAQLARDLWAGGLRAGLHAGDAPDLAPGAVLAEHDPPLVRLSCWDQDRIREHRVPYVDVVDFIKQKIIGDSSRNPEGSSTNRSISWSESSEKYNSPNISVTFITASDRMTKNSKRHCENQINTQVTSILVSLGLLPVMARVRVNVLAVACDSACVRYMASQLTAPLDADDLSQAFRNVTEMFPKRQGLIDEARSELSSLLKQTSPSRNNEETQIYALYSIPDSLCRLIT
Summary
Description
Metabolic-stress sensing protein kinase that phosphorylates the alpha subunit of eukaryotic translation initiation factor 2 (eIF-2-alpha/EIF2S1) on 'Ser-52' in response to low amino acid availability (PubMed:10504407, PubMed:10655230, PubMed:12176355, PubMed:12215525, PubMed:15213227, PubMed:16054071, PubMed:16176978, PubMed:16121183, PubMed:15774759, PubMed:16601681, PubMed:26102367). Plays a role as an activator of the integrated stress response (ISR) required for adapatation to amino acid starvation. Converts phosphorylated eIF-2-alpha/EIF2S1 either to a competitive inhibitor of the translation initiation factor eIF-2B, leading to a global protein synthesis repression, and thus to a reduced overall utilization of amino acids, or to a translational initiation activation of specific mRNAs, such as the transcriptional activator ATF4, and hence allowing ATF4-mediated reprogramming of amino acid biosynthetic gene expression to alleviate nutrient depletion (PubMed:10655230, PubMed:11106749, PubMed:12176355, PubMed:15213227, PubMed:16176978, PubMed:26102367). Binds uncharged tRNAs (By similarity). Involved in cell cycle arrest by promoting cyclin D1 mRNA translation repression after the unfolded protein response pathway (UPR) activation or cell cycle inhibitor CDKN1A/p21 mRNA translation activation in response to amino acid deprivation (PubMed:16176978, PubMed:26102367). Plays a role in the consolidation of synaptic plasticity, learning as well as formation of long-term memory (PubMed:16121183). Plays a role in neurite outgrowth inhibition (PubMed:23447528). Plays a role in feeding behavior to maintain amino acid homeostasis; contributes to the innate aversion toward diets of imbalanced amino acid composition (PubMed:16054071, PubMed:15774759). Plays a proapoptotic role in response to glucose deprivation (PubMed:20660158). Promotes global cellular protein synthesis repression in response to UV irradiation independently of the stress-activated protein kinase/c-Jun N-terminal kinase (SAPK/JNK) and p38 MAPK signaling pathways (PubMed:12176355).
(Microbial infection) Plays a role in the antiviral response against alphavirus infection; impairs early viral mRNA translation of the incoming genomic virus RNA, thus preventing alphavirus replication.
(Microbial infection) Plays a role in modulating the adaptive immune response to Yellow fever virus infection; promotes dendritic cells to initiate autophagy and antigene presentation to both CD4(+) and CD8(+) T-cells under amino acid starvation.
Metabolic-stress sensing protein kinase that phosphorylates the alpha subunit of eukaryotic translation initiation factor 2 (eIF-2-alpha/EIF2S1) on 'Ser-52' in response to low amino acid availability (PubMed:25329545). Plays a role as an activator of the integrated stress response (ISR) required for adapatation to amino acid starvation. Converts phosphorylated eIF-2-alpha/EIF2S1 either to a competitive inhibitor of the translation initiation factor eIF-2B, leading to a global protein synthesis repression, and thus to a reduced overall utilization of amino acids, or to a translational initiation activation of specific mRNAs, such as the transcriptional activator ATF4, and hence allowing ATF4-mediated reprogramming of amino acid biosynthetic gene expression to alleviate nutrient depletion. Binds uncharged tRNAs (By similarity). Involved in cell cycle arrest by promoting cyclin D1 mRNA translation repression after the unfolded protein response pathway (UPR) activation or cell cycle inhibitor CDKN1A/p21 mRNA translation activation in response to amino acid deprivation (PubMed:26102367). Plays a role in the consolidation of synaptic plasticity, learning as well as formation of long-term memory. Plays a role in neurite outgrowth inhibition. Plays a proapoptotic role in response to glucose deprivation. Promotes global cellular protein synthesis repression in response to UV irradiation independently of the stress-activated protein kinase/c-Jun N-terminal kinase (SAPK/JNK) and p38 MAPK signaling pathways (By similarity). Plays a role in the antiviral response against alphavirus infection; impairs early viral mRNA translation of the incoming genomic virus RNA, thus preventing alphavirus replication (By similarity).
(Microbial infection) Plays a role in modulating the adaptive immune response to yellow fever virus infection; promotes dendritic cells to initiate autophagy and antigene presentation to both CD4(+) and CD8(+) T-cells under amino acid starvation (PubMed:24310610).
(Microbial infection) Plays a role in the antiviral response against alphavirus infection; impairs early viral mRNA translation of the incoming genomic virus RNA, thus preventing alphavirus replication.
(Microbial infection) Plays a role in modulating the adaptive immune response to Yellow fever virus infection; promotes dendritic cells to initiate autophagy and antigene presentation to both CD4(+) and CD8(+) T-cells under amino acid starvation.
Metabolic-stress sensing protein kinase that phosphorylates the alpha subunit of eukaryotic translation initiation factor 2 (eIF-2-alpha/EIF2S1) on 'Ser-52' in response to low amino acid availability (PubMed:25329545). Plays a role as an activator of the integrated stress response (ISR) required for adapatation to amino acid starvation. Converts phosphorylated eIF-2-alpha/EIF2S1 either to a competitive inhibitor of the translation initiation factor eIF-2B, leading to a global protein synthesis repression, and thus to a reduced overall utilization of amino acids, or to a translational initiation activation of specific mRNAs, such as the transcriptional activator ATF4, and hence allowing ATF4-mediated reprogramming of amino acid biosynthetic gene expression to alleviate nutrient depletion. Binds uncharged tRNAs (By similarity). Involved in cell cycle arrest by promoting cyclin D1 mRNA translation repression after the unfolded protein response pathway (UPR) activation or cell cycle inhibitor CDKN1A/p21 mRNA translation activation in response to amino acid deprivation (PubMed:26102367). Plays a role in the consolidation of synaptic plasticity, learning as well as formation of long-term memory. Plays a role in neurite outgrowth inhibition. Plays a proapoptotic role in response to glucose deprivation. Promotes global cellular protein synthesis repression in response to UV irradiation independently of the stress-activated protein kinase/c-Jun N-terminal kinase (SAPK/JNK) and p38 MAPK signaling pathways (By similarity). Plays a role in the antiviral response against alphavirus infection; impairs early viral mRNA translation of the incoming genomic virus RNA, thus preventing alphavirus replication (By similarity).
(Microbial infection) Plays a role in modulating the adaptive immune response to yellow fever virus infection; promotes dendritic cells to initiate autophagy and antigene presentation to both CD4(+) and CD8(+) T-cells under amino acid starvation (PubMed:24310610).
Catalytic Activity
ATP + L-seryl-[protein] = ADP + H(+) + O-phospho-L-seryl-[protein]
ATP + L-threonyl-[protein] = ADP + H(+) + O-phospho-L-threonyl-[protein]
ATP + L-threonyl-[protein] = ADP + H(+) + O-phospho-L-threonyl-[protein]
Subunit
Homodimer; homodimerization is important for kinase activation by uncharged tRNAs (By similarity). Interacts with GCN1; this interaction stimulates EIF2AK4/GCN2 kinase activity and is impaired by IMPACT upon a variety of stress conditions, such as amino acid depletion, UV-C irradiation, proteasome inhibitor treatment and glucose deprivation (PubMed:24333428). Interacts with DNAJC3; this interaction inhibits EIF2AK4/GCN2 kinase activity during endoplasmic reticulum (ER), hypothermic and amino acid-starving stress conditions (PubMed:25329545).
Homodimer; homodimerization is important for kinase activation by uncharged tRNAs. Interacts with GCN1; this interaction stimulates EIF2AK4/GCN2 kinase activity and is impaired by IMPACT upon a variety of stress conditions, such as amino acid depletion, UV-C irradiation, proteasome inhibitor treatment and glucose deprivation. Interacts with DNAJC3; this interaction inhibits EIF2AK4/GCN2 kinase activity during endoplasmic reticulum (ER), hypothermic and amino acid-starving stress conditions.
Homodimer; homodimerization is important for kinase activation by uncharged tRNAs. Interacts with GCN1; this interaction stimulates EIF2AK4/GCN2 kinase activity and is impaired by IMPACT upon a variety of stress conditions, such as amino acid depletion, UV-C irradiation, proteasome inhibitor treatment and glucose deprivation. Interacts with DNAJC3; this interaction inhibits EIF2AK4/GCN2 kinase activity during endoplasmic reticulum (ER), hypothermic and amino acid-starving stress conditions.
Similarity
Belongs to the protein kinase superfamily. Tyr protein kinase family.
Belongs to the protein kinase superfamily. Ser/Thr protein kinase family. GCN2 subfamily.
Belongs to the protein kinase superfamily. Ser/Thr protein kinase family. GCN2 subfamily.
Keywords
3D-structure
Acetylation
Activation of host autophagy by virus
Activator
Adaptive immunity
Alternative splicing
Antiviral defense
ATP-binding
Cell cycle
Coiled coil
Complete proteome
Cytoplasm
Differentiation
Direct protein sequencing
Growth arrest
Host-virus interaction
Immunity
Kinase
Neurogenesis
Nucleotide-binding
Phosphoprotein
Reference proteome
Repeat
RNA-binding
Stress response
Transferase
Translation regulation
tRNA-binding
Disease mutation
Polymorphism
Feature
chain eIF-2-alpha kinase GCN2
splice variant In isoform 5.
sequence variant In dbSNP:rs35509999.
splice variant In isoform 5.
sequence variant In dbSNP:rs35509999.
Uniprot
H9J5T2
A0A2A4JSE2
A0A212FG33
A0A2H1V9T6
A0A2W1BXS8
A0A3S2LU79
+ More
A0A232ESJ4 A0A151J0K5 A0A151I1P2 A0A158NAF7 A0A1Y1LNY7 A0A0C9QMU9 D6WSZ3 A0A0C9QMD1 A0A2J7Q260 T1IQL0 A0A088AEP3 A0A1I8NAC2 A0A1L8FAK5 A0A1Y1LP48 F7BV00 A0A1Q3F239 F7BH85 I3K2D0 A0A0M3QYL7 M3X137 G7MWM5 A0A096NTE4 A0A2K5NYZ9 A0A0D9R4Q6 A0A2K5ZJC5 A0A2J8T2Y6 H9GL93 A0A2K6NN09 A0A3L8SY72 A0A2K5E3L8 A0A3Q3C620 G3S852 A0A2R9AQ16 A0A2K5K4A5 Q9QZ05 Q9P2K8 H2Q956 A2AUM0 A0A2R9AQ06 E9G227 Q4RLP9 A0A3B3RFP2 U3FQ30 O01712 A0A218V8Q9 U3BNN6 A0A3B3RE15 Q9V9X8 L5K1I4 O61651 B4I024 Q9P2K8-2 H0ZMZ5 A0A3B4U648 A0A2J8QDB7 A0A2D0RNG7 A0A0B4KHX7 A0A0P5YXY2 A0A1S3AMM2 A0A1A7YRD5 A0A0N8A753 A0A0P5VZ04 A0A0P5AFM3 A0A0P4YHK3 A0A0P5U427 K7FQP3 A0A0N8D0E0 A0A286XMJ2 A0A0P4Y5R0 A0A087V2U7 A0A0P5XMK4 A0A3P9Q7K0 A0A0P5ZNA9 A0A091MTS3 A0A0P5X1S8 A0A3P9K4E3 A0A3Q2UGU9 A0A0N8EQA6 M3ZXB5 A0A1A7XTG5 A0A3Q2DB77 A0A3Q1BQ84 A0A091IWZ4 A0A0P5CMS7 A0A2C9KLS5 A0A2U9CJ75 I3MQ20 A0A146V5I0 A0A087XYY3 A0A0N7ZU46 A0A0P5D2A7 A0A146MR45 A0A2I0MP31 A0A3B3VZ94
A0A232ESJ4 A0A151J0K5 A0A151I1P2 A0A158NAF7 A0A1Y1LNY7 A0A0C9QMU9 D6WSZ3 A0A0C9QMD1 A0A2J7Q260 T1IQL0 A0A088AEP3 A0A1I8NAC2 A0A1L8FAK5 A0A1Y1LP48 F7BV00 A0A1Q3F239 F7BH85 I3K2D0 A0A0M3QYL7 M3X137 G7MWM5 A0A096NTE4 A0A2K5NYZ9 A0A0D9R4Q6 A0A2K5ZJC5 A0A2J8T2Y6 H9GL93 A0A2K6NN09 A0A3L8SY72 A0A2K5E3L8 A0A3Q3C620 G3S852 A0A2R9AQ16 A0A2K5K4A5 Q9QZ05 Q9P2K8 H2Q956 A2AUM0 A0A2R9AQ06 E9G227 Q4RLP9 A0A3B3RFP2 U3FQ30 O01712 A0A218V8Q9 U3BNN6 A0A3B3RE15 Q9V9X8 L5K1I4 O61651 B4I024 Q9P2K8-2 H0ZMZ5 A0A3B4U648 A0A2J8QDB7 A0A2D0RNG7 A0A0B4KHX7 A0A0P5YXY2 A0A1S3AMM2 A0A1A7YRD5 A0A0N8A753 A0A0P5VZ04 A0A0P5AFM3 A0A0P4YHK3 A0A0P5U427 K7FQP3 A0A0N8D0E0 A0A286XMJ2 A0A0P4Y5R0 A0A087V2U7 A0A0P5XMK4 A0A3P9Q7K0 A0A0P5ZNA9 A0A091MTS3 A0A0P5X1S8 A0A3P9K4E3 A0A3Q2UGU9 A0A0N8EQA6 M3ZXB5 A0A1A7XTG5 A0A3Q2DB77 A0A3Q1BQ84 A0A091IWZ4 A0A0P5CMS7 A0A2C9KLS5 A0A2U9CJ75 I3MQ20 A0A146V5I0 A0A087XYY3 A0A0N7ZU46 A0A0P5D2A7 A0A146MR45 A0A2I0MP31 A0A3B3VZ94
EC Number
2.7.11.1
Pubmed
19121390
22118469
28756777
28648823
21347285
28004739
+ More
18362917 19820115 25315136 27762356 20431018 25186727 17975172 22002653 21881562 25362486 30282656 22398555 22722832 10504407 10655230 16141072 19468303 15489334 14621295 11106749 12176355 12215525 15213227 16054071 16176978 16121183 15774759 16601681 21183079 20660158 23447528 24333428 24310610 25329545 26102367 15273307 16572171 10718198 17974005 14702039 17081983 18691976 18669648 19369195 19608861 20068231 21269460 21406692 23186163 24135949 24292273 17344846 16136131 21292972 15496914 29240929 25243066 9139706 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 23258410 9649537 17994087 20360741 17381049 21993624 17554307 23542700 15562597 23371554
18362917 19820115 25315136 27762356 20431018 25186727 17975172 22002653 21881562 25362486 30282656 22398555 22722832 10504407 10655230 16141072 19468303 15489334 14621295 11106749 12176355 12215525 15213227 16054071 16176978 16121183 15774759 16601681 21183079 20660158 23447528 24333428 24310610 25329545 26102367 15273307 16572171 10718198 17974005 14702039 17081983 18691976 18669648 19369195 19608861 20068231 21269460 21406692 23186163 24135949 24292273 17344846 16136131 21292972 15496914 29240929 25243066 9139706 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 23258410 9649537 17994087 20360741 17381049 21993624 17554307 23542700 15562597 23371554
EMBL
BABH01025161
BABH01025162
BABH01025163
BABH01025164
NWSH01000660
PCG74957.1
+ More
AGBW02008725 OWR52683.1 ODYU01001427 SOQ37598.1 KZ149903 PZC78444.1 RSAL01000009 RVE53876.1 NNAY01002415 OXU21328.1 KQ980629 KYN15001.1 KQ976600 KYM79468.1 ADTU01010273 GEZM01050768 JAV75394.1 GBYB01001982 JAG71749.1 KQ971352 EFA06330.1 GBYB01004674 JAG74441.1 NEVH01019373 PNF22671.1 JH431312 CM004480 OCT68625.1 GEZM01050759 JAV75409.1 AAMC01063663 AAMC01063664 AAMC01063665 GFDL01013422 JAV21623.1 AERX01018652 AERX01018653 CP012526 ALC47839.1 AANG04001761 CM001259 EHH27200.1 AHZZ02030793 AHZZ02030794 AQIB01116350 AQIB01116351 AQIB01116352 NDHI03003524 PNJ27389.1 AAWZ02022551 QUSF01000004 RLW10462.1 CABD030095524 AJFE02093060 AJFE02093061 AJFE02093062 AJFE02093063 AJ243533 AF193342 AF193343 AF193344 AK011380 AK013758 AK077199 AL929163 BC023958 BC072637 AK129334 AC012377 AC025168 BC009350 BC078179 AB037759 AL137627 AL157497 AL832907 AK027011 AJ243428 AACZ04038090 AACZ04038091 GABC01006885 GABC01006884 GABC01006883 GABC01006882 GABC01006881 GABD01001668 GABD01001667 GABD01001666 GABD01001665 GABE01005382 GABE01005381 GABE01005380 GABE01005379 GABE01005378 GABE01005377 GABE01005376 NBAG03000046 JAA04453.1 JAA31432.1 JAA39357.1 PNI94271.1 GL732530 EFX86379.1 CAAE01015019 CAG10683.1 GAMS01003843 GAMS01003842 GAMR01003306 GAMP01004630 GAMP01004629 JAB19293.1 JAB30626.1 JAB48126.1 U80223 AAC47516.1 MUZQ01000033 OWK61992.1 GAMT01004457 JAB07404.1 AE014297 BT126024 AAF57150.1 ADY17723.1 AGB96520.1 KB031042 ELK05554.1 AF056302 AAC13490.1 CH480819 EDW53631.1 ABQF01002671 PNI94270.1 AGB96521.1 GDIP01052237 JAM51478.1 HADX01010885 SBP33117.1 GDIP01175913 JAJ47489.1 GDIP01107721 LRGB01002384 JAL95993.1 KZS07872.1 GDIP01199861 JAJ23541.1 GDIP01230861 JAI92540.1 GDIP01118422 JAL85292.1 AGCU01123582 AGCU01123583 AGCU01123584 AGCU01123585 AGCU01123586 AGCU01123587 AGCU01123588 AGCU01123589 AGCU01123590 GDIP01081127 JAM22588.1 AAKN02015867 AAKN02015868 GDIP01232108 JAI91293.1 KL477821 KFO06939.1 GDIP01069926 JAM33789.1 GDIP01041532 JAM62183.1 KL371374 KFP80691.1 GDIP01078626 JAM25089.1 GDIQ01004420 JAN90317.1 HADW01019971 SBP21371.1 KK501012 KFP12093.1 GDIP01168263 JAJ55139.1 CP026259 AWP16645.1 AGTP01002143 AGTP01002144 AGTP01002145 AGTP01002146 AGTP01002147 AGTP01002148 AGTP01002149 AGTP01002150 GCES01073633 JAR12690.1 AYCK01003405 GDIP01212369 JAJ11033.1 GDIP01167415 JAJ55987.1 GCES01165192 JAQ21130.1 AKCR02000005 PKK31430.1
AGBW02008725 OWR52683.1 ODYU01001427 SOQ37598.1 KZ149903 PZC78444.1 RSAL01000009 RVE53876.1 NNAY01002415 OXU21328.1 KQ980629 KYN15001.1 KQ976600 KYM79468.1 ADTU01010273 GEZM01050768 JAV75394.1 GBYB01001982 JAG71749.1 KQ971352 EFA06330.1 GBYB01004674 JAG74441.1 NEVH01019373 PNF22671.1 JH431312 CM004480 OCT68625.1 GEZM01050759 JAV75409.1 AAMC01063663 AAMC01063664 AAMC01063665 GFDL01013422 JAV21623.1 AERX01018652 AERX01018653 CP012526 ALC47839.1 AANG04001761 CM001259 EHH27200.1 AHZZ02030793 AHZZ02030794 AQIB01116350 AQIB01116351 AQIB01116352 NDHI03003524 PNJ27389.1 AAWZ02022551 QUSF01000004 RLW10462.1 CABD030095524 AJFE02093060 AJFE02093061 AJFE02093062 AJFE02093063 AJ243533 AF193342 AF193343 AF193344 AK011380 AK013758 AK077199 AL929163 BC023958 BC072637 AK129334 AC012377 AC025168 BC009350 BC078179 AB037759 AL137627 AL157497 AL832907 AK027011 AJ243428 AACZ04038090 AACZ04038091 GABC01006885 GABC01006884 GABC01006883 GABC01006882 GABC01006881 GABD01001668 GABD01001667 GABD01001666 GABD01001665 GABE01005382 GABE01005381 GABE01005380 GABE01005379 GABE01005378 GABE01005377 GABE01005376 NBAG03000046 JAA04453.1 JAA31432.1 JAA39357.1 PNI94271.1 GL732530 EFX86379.1 CAAE01015019 CAG10683.1 GAMS01003843 GAMS01003842 GAMR01003306 GAMP01004630 GAMP01004629 JAB19293.1 JAB30626.1 JAB48126.1 U80223 AAC47516.1 MUZQ01000033 OWK61992.1 GAMT01004457 JAB07404.1 AE014297 BT126024 AAF57150.1 ADY17723.1 AGB96520.1 KB031042 ELK05554.1 AF056302 AAC13490.1 CH480819 EDW53631.1 ABQF01002671 PNI94270.1 AGB96521.1 GDIP01052237 JAM51478.1 HADX01010885 SBP33117.1 GDIP01175913 JAJ47489.1 GDIP01107721 LRGB01002384 JAL95993.1 KZS07872.1 GDIP01199861 JAJ23541.1 GDIP01230861 JAI92540.1 GDIP01118422 JAL85292.1 AGCU01123582 AGCU01123583 AGCU01123584 AGCU01123585 AGCU01123586 AGCU01123587 AGCU01123588 AGCU01123589 AGCU01123590 GDIP01081127 JAM22588.1 AAKN02015867 AAKN02015868 GDIP01232108 JAI91293.1 KL477821 KFO06939.1 GDIP01069926 JAM33789.1 GDIP01041532 JAM62183.1 KL371374 KFP80691.1 GDIP01078626 JAM25089.1 GDIQ01004420 JAN90317.1 HADW01019971 SBP21371.1 KK501012 KFP12093.1 GDIP01168263 JAJ55139.1 CP026259 AWP16645.1 AGTP01002143 AGTP01002144 AGTP01002145 AGTP01002146 AGTP01002147 AGTP01002148 AGTP01002149 AGTP01002150 GCES01073633 JAR12690.1 AYCK01003405 GDIP01212369 JAJ11033.1 GDIP01167415 JAJ55987.1 GCES01165192 JAQ21130.1 AKCR02000005 PKK31430.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000283053
UP000215335
UP000078492
+ More
UP000078540 UP000005205 UP000007266 UP000235965 UP000005203 UP000095301 UP000186698 UP000008143 UP000005207 UP000092553 UP000011712 UP000028761 UP000233060 UP000029965 UP000233140 UP000001646 UP000233200 UP000276834 UP000233020 UP000264840 UP000001519 UP000240080 UP000233080 UP000000589 UP000005640 UP000002277 UP000000305 UP000261540 UP000008225 UP000197619 UP000000803 UP000010552 UP000001292 UP000007754 UP000261420 UP000221080 UP000079721 UP000076858 UP000007267 UP000005447 UP000242638 UP000265180 UP000265000 UP000002852 UP000265020 UP000257160 UP000053119 UP000076420 UP000246464 UP000005215 UP000028760 UP000053872 UP000261500
UP000078540 UP000005205 UP000007266 UP000235965 UP000005203 UP000095301 UP000186698 UP000008143 UP000005207 UP000092553 UP000011712 UP000028761 UP000233060 UP000029965 UP000233140 UP000001646 UP000233200 UP000276834 UP000233020 UP000264840 UP000001519 UP000240080 UP000233080 UP000000589 UP000005640 UP000002277 UP000000305 UP000261540 UP000008225 UP000197619 UP000000803 UP000010552 UP000001292 UP000007754 UP000261420 UP000221080 UP000079721 UP000076858 UP000007267 UP000005447 UP000242638 UP000265180 UP000265000 UP000002852 UP000265020 UP000257160 UP000053119 UP000076420 UP000246464 UP000005215 UP000028760 UP000053872 UP000261500
Interpro
IPR041715
HisRS-like_core
+ More
IPR020635 Tyr_kinase_cat_dom
IPR016135 UBQ-conjugating_enzyme/RWD
IPR000719 Prot_kinase_dom
IPR011009 Kinase-like_dom_sf
IPR006575 RWD-domain
IPR016255 Gcn2
IPR008266 Tyr_kinase_AS
IPR008271 Ser/Thr_kinase_AS
IPR017441 Protein_kinase_ATP_BS
IPR036621 Anticodon-bd_dom_sf
IPR001611 Leu-rich_rpt
IPR024435 HisRS-related_dom
IPR006195 aa-tRNA-synth_II
IPR020635 Tyr_kinase_cat_dom
IPR016135 UBQ-conjugating_enzyme/RWD
IPR000719 Prot_kinase_dom
IPR011009 Kinase-like_dom_sf
IPR006575 RWD-domain
IPR016255 Gcn2
IPR008266 Tyr_kinase_AS
IPR008271 Ser/Thr_kinase_AS
IPR017441 Protein_kinase_ATP_BS
IPR036621 Anticodon-bd_dom_sf
IPR001611 Leu-rich_rpt
IPR024435 HisRS-related_dom
IPR006195 aa-tRNA-synth_II
Gene 3D
ProteinModelPortal
H9J5T2
A0A2A4JSE2
A0A212FG33
A0A2H1V9T6
A0A2W1BXS8
A0A3S2LU79
+ More
A0A232ESJ4 A0A151J0K5 A0A151I1P2 A0A158NAF7 A0A1Y1LNY7 A0A0C9QMU9 D6WSZ3 A0A0C9QMD1 A0A2J7Q260 T1IQL0 A0A088AEP3 A0A1I8NAC2 A0A1L8FAK5 A0A1Y1LP48 F7BV00 A0A1Q3F239 F7BH85 I3K2D0 A0A0M3QYL7 M3X137 G7MWM5 A0A096NTE4 A0A2K5NYZ9 A0A0D9R4Q6 A0A2K5ZJC5 A0A2J8T2Y6 H9GL93 A0A2K6NN09 A0A3L8SY72 A0A2K5E3L8 A0A3Q3C620 G3S852 A0A2R9AQ16 A0A2K5K4A5 Q9QZ05 Q9P2K8 H2Q956 A2AUM0 A0A2R9AQ06 E9G227 Q4RLP9 A0A3B3RFP2 U3FQ30 O01712 A0A218V8Q9 U3BNN6 A0A3B3RE15 Q9V9X8 L5K1I4 O61651 B4I024 Q9P2K8-2 H0ZMZ5 A0A3B4U648 A0A2J8QDB7 A0A2D0RNG7 A0A0B4KHX7 A0A0P5YXY2 A0A1S3AMM2 A0A1A7YRD5 A0A0N8A753 A0A0P5VZ04 A0A0P5AFM3 A0A0P4YHK3 A0A0P5U427 K7FQP3 A0A0N8D0E0 A0A286XMJ2 A0A0P4Y5R0 A0A087V2U7 A0A0P5XMK4 A0A3P9Q7K0 A0A0P5ZNA9 A0A091MTS3 A0A0P5X1S8 A0A3P9K4E3 A0A3Q2UGU9 A0A0N8EQA6 M3ZXB5 A0A1A7XTG5 A0A3Q2DB77 A0A3Q1BQ84 A0A091IWZ4 A0A0P5CMS7 A0A2C9KLS5 A0A2U9CJ75 I3MQ20 A0A146V5I0 A0A087XYY3 A0A0N7ZU46 A0A0P5D2A7 A0A146MR45 A0A2I0MP31 A0A3B3VZ94
A0A232ESJ4 A0A151J0K5 A0A151I1P2 A0A158NAF7 A0A1Y1LNY7 A0A0C9QMU9 D6WSZ3 A0A0C9QMD1 A0A2J7Q260 T1IQL0 A0A088AEP3 A0A1I8NAC2 A0A1L8FAK5 A0A1Y1LP48 F7BV00 A0A1Q3F239 F7BH85 I3K2D0 A0A0M3QYL7 M3X137 G7MWM5 A0A096NTE4 A0A2K5NYZ9 A0A0D9R4Q6 A0A2K5ZJC5 A0A2J8T2Y6 H9GL93 A0A2K6NN09 A0A3L8SY72 A0A2K5E3L8 A0A3Q3C620 G3S852 A0A2R9AQ16 A0A2K5K4A5 Q9QZ05 Q9P2K8 H2Q956 A2AUM0 A0A2R9AQ06 E9G227 Q4RLP9 A0A3B3RFP2 U3FQ30 O01712 A0A218V8Q9 U3BNN6 A0A3B3RE15 Q9V9X8 L5K1I4 O61651 B4I024 Q9P2K8-2 H0ZMZ5 A0A3B4U648 A0A2J8QDB7 A0A2D0RNG7 A0A0B4KHX7 A0A0P5YXY2 A0A1S3AMM2 A0A1A7YRD5 A0A0N8A753 A0A0P5VZ04 A0A0P5AFM3 A0A0P4YHK3 A0A0P5U427 K7FQP3 A0A0N8D0E0 A0A286XMJ2 A0A0P4Y5R0 A0A087V2U7 A0A0P5XMK4 A0A3P9Q7K0 A0A0P5ZNA9 A0A091MTS3 A0A0P5X1S8 A0A3P9K4E3 A0A3Q2UGU9 A0A0N8EQA6 M3ZXB5 A0A1A7XTG5 A0A3Q2DB77 A0A3Q1BQ84 A0A091IWZ4 A0A0P5CMS7 A0A2C9KLS5 A0A2U9CJ75 I3MQ20 A0A146V5I0 A0A087XYY3 A0A0N7ZU46 A0A0P5D2A7 A0A146MR45 A0A2I0MP31 A0A3B3VZ94
PDB
1ZYD
E-value=4.18024e-42,
Score=437
Ontologies
PATHWAY
GO
GO:0000077
GO:0004713
GO:0004694
GO:0005524
GO:0003743
GO:0004672
GO:0022626
GO:0036492
GO:0060733
GO:0070417
GO:0006446
GO:1990138
GO:0010628
GO:0007050
GO:0032057
GO:0019081
GO:0007612
GO:0060259
GO:1990253
GO:0046777
GO:0030968
GO:0017148
GO:0006468
GO:0010998
GO:0002286
GO:0071264
GO:1900273
GO:0002250
GO:0045665
GO:0034644
GO:0032792
GO:0044828
GO:0004674
GO:0007616
GO:0034198
GO:0005844
GO:0009267
GO:0039520
GO:0034976
GO:0045947
GO:0043558
GO:0051607
GO:0002230
GO:0000049
GO:0071074
GO:0002821
GO:0045793
GO:0004686
GO:0005829
GO:0005515
GO:0003676
GO:0007154
GO:0016020
GO:0005112
GO:0015930
Topology
Subcellular location
Cytoplasm
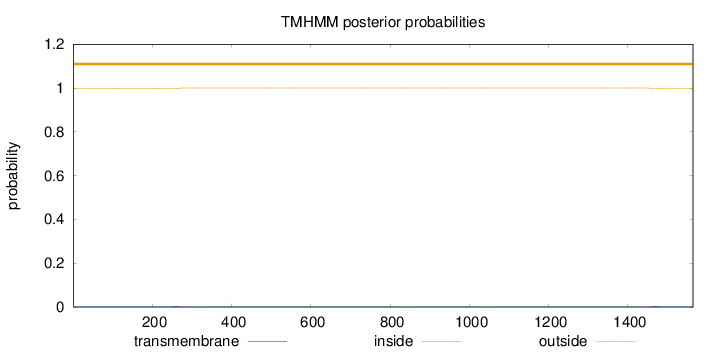
Length:
1564
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.08324
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00157
outside
1 - 1564
Population Genetic Test Statistics
Pi
187.810526
Theta
163.054302
Tajima's D
0.693894
CLR
0.280569
CSRT
0.574621268936553
Interpretation
Uncertain
