Gene
KWMTBOMO15010
Pre Gene Modal
BGIBMGA004795
Annotation
PREDICTED:_sodium/calcium_exchanger_2_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.169
Sequence
CDS
ATGATAGTCTCTCTCCATTTCCAAAAGTCCCTTACCATCGATAACGATGATTCGAATACTGGTACCGAAGCCAACCATCTCGCAACGCTGCCGGAAGAGGAAGCAGTAGTCGCCATGATGGGACTCCCTAAAGCCGGGGAAATCACACGAGCGCAGCTTAGGATCAAGGAGAGCAAAGAGTTTAAGAACACAGTAGACAAATTGGTGCAAAGAGCGAACGCGTCTATAATCATAGGAACGTCTTCATGGAAGGAGCAATTCGCGGAAGCCTTAACCGCCAATTCAGGTTCAGACGACCCAGACGAGCCTCCTTCGTGCTTTGACTACGTTCTCCACGTGGTCACTTTGTTCTGGAAAATCATATTCGCTTTGATACCACCGACAGACATCGGCGGTGGCTACGTCTGCTTTGTGGTTTCGATAATTTGCATCGGCCTGGTGACGGCTGTGATTGGAGATGTCGCGTCACACTTCGGCTGTACTCTTGGCATCAAGGATTCTGTCACAGCCATCGTATTTGTGGCTTTGGGCACGAGTATTCCTGACACGTTTGCGAGTAAAGTGGCCGCTATCCAAGATAAGCACGCGGATGCTTCTGTTGGCAACGTGACCGGCTCTAATGCTGTGAACGTATTCCTTGGTATCGGCGTCGCGTGGACGGTGGCCGCCATCTTCCACTGGAGTAAGAACGAGAAGTTCCACGTGGACCCTGGGAACCTAGCCTTCAGCGTCACGATGTTCTGCTCGGAAGCCGCTCTAGCTGTCCTTGTCCTGGTGCTGAGAAGACGTCGAAGTATTGGCGGGGAGCTCGGCGGACCACGCGGCATCAAGATCGTAACTTCCATGTTCTTCTTTTCCCTGTGGCTAATCTATCTAATCATGTCTGGCTTGGAAGCCTATGACGTCATAGAAGGTTTTTGA
Protein
MIVSLHFQKSLTIDNDDSNTGTEANHLATLPEEEAVVAMMGLPKAGEITRAQLRIKESKEFKNTVDKLVQRANASIIIGTSSWKEQFAEALTANSGSDDPDEPPSCFDYVLHVVTLFWKIIFALIPPTDIGGGYVCFVVSIICIGLVTAVIGDVASHFGCTLGIKDSVTAIVFVALGTSIPDTFASKVAAIQDKHADASVGNVTGSNAVNVFLGIGVAWTVAAIFHWSKNEKFHVDPGNLAFSVTMFCSEAALAVLVLVLRRRRSIGGELGGPRGIKIVTSMFFFSLWLIYLIMSGLEAYDVIEGF
Summary
Similarity
Belongs to the Ca(2+):cation antiporter (CaCA) (TC 2.A.19) family.
Uniprot
H9J5K5
A0A2A4K7T9
A0A2W1BTN1
A0A3S2TSG4
A0A194PLS1
A0A0L7LDH1
+ More
E9J4G5 A0A340TBD3 A0A1L8DFA0 A0A1Q3FVG2 A0A1Q3FVJ1 E2BQW7 A0A1Y1NDC9 F4X5U0 E2A904 N6TK22 A0A1I8Q2V1 A0A151ISW7 U4UFD0 A0A1W4WH66 A0A1W4W6T8 A0A1A9YLF1 A0A0K8W5L6 A0A1B0BG51 A0A3L8DJC3 A0A026WFF4 A0A195D775 A0A0A1XAD6 A0A1Y1NCB8 A0A151WVJ6 A0A158NME2 A0A151JX34 A0A151I3A1 D6WRH9 T1PCS8 T1PL58 W8AQ93 W8AL91 A0A1I8M5B8 B0WFB5 A0A067RIB9 A0A182K3A9 A0A182T391 A0A1B0GG32 Q7QCY1 A0A182RQ24 A0A154P3C3 A0A182LGC3 K7J312 A0A0C9QG73 K7J313 A0A182XHL8 A0A182HTS4 A0A182TX31 A0A182R083 V9IHT9 A0A182J9S9 A0A1B0CRK4 A0A194RHJ1 A0A182FR16 A0A0A1WL41 A0A0L7QTV0 A0A034VRI2 A0A034VUX1 A0A034VVP4 A0A034VVP9 A0A182VDX8 A0A088AB77 A0A084VDR1 A0A1J1IYD1 A0A1S3PGE9 A0A3B4VL73 A0A060VZ30 A0A1S3L856 A0A3B4VNN4 A0A3B4VJY0 A0A1A9VQY0 A0A3P9NCJ6 A0A1I8NE62 A0A3P9NCG5 A0A060W262 A0A0S7L156 A0A3N0YNZ1 A0A3B5PYK3 M4AMU3 A0A3B5RB01 A0A182XW68 A0A3B5PRY8 A0A1A8S554 B7QAE5 A0A3B5LSV5 A0A0F8CHZ0 A0A286YBI0 F6PC65 F1R4F4 Q32SG8 A0A023GL62 G3Q8Q6 G3Q8Q8 G3Q8Q4
E9J4G5 A0A340TBD3 A0A1L8DFA0 A0A1Q3FVG2 A0A1Q3FVJ1 E2BQW7 A0A1Y1NDC9 F4X5U0 E2A904 N6TK22 A0A1I8Q2V1 A0A151ISW7 U4UFD0 A0A1W4WH66 A0A1W4W6T8 A0A1A9YLF1 A0A0K8W5L6 A0A1B0BG51 A0A3L8DJC3 A0A026WFF4 A0A195D775 A0A0A1XAD6 A0A1Y1NCB8 A0A151WVJ6 A0A158NME2 A0A151JX34 A0A151I3A1 D6WRH9 T1PCS8 T1PL58 W8AQ93 W8AL91 A0A1I8M5B8 B0WFB5 A0A067RIB9 A0A182K3A9 A0A182T391 A0A1B0GG32 Q7QCY1 A0A182RQ24 A0A154P3C3 A0A182LGC3 K7J312 A0A0C9QG73 K7J313 A0A182XHL8 A0A182HTS4 A0A182TX31 A0A182R083 V9IHT9 A0A182J9S9 A0A1B0CRK4 A0A194RHJ1 A0A182FR16 A0A0A1WL41 A0A0L7QTV0 A0A034VRI2 A0A034VUX1 A0A034VVP4 A0A034VVP9 A0A182VDX8 A0A088AB77 A0A084VDR1 A0A1J1IYD1 A0A1S3PGE9 A0A3B4VL73 A0A060VZ30 A0A1S3L856 A0A3B4VNN4 A0A3B4VJY0 A0A1A9VQY0 A0A3P9NCJ6 A0A1I8NE62 A0A3P9NCG5 A0A060W262 A0A0S7L156 A0A3N0YNZ1 A0A3B5PYK3 M4AMU3 A0A3B5RB01 A0A182XW68 A0A3B5PRY8 A0A1A8S554 B7QAE5 A0A3B5LSV5 A0A0F8CHZ0 A0A286YBI0 F6PC65 F1R4F4 Q32SG8 A0A023GL62 G3Q8Q6 G3Q8Q8 G3Q8Q4
Pubmed
EMBL
BABH01025171
BABH01025172
NWSH01000067
PCG79998.1
KZ149903
PZC78442.1
+ More
RSAL01000009 RVE53873.1 KQ459604 KPI92055.1 JTDY01001560 KOB73527.1 GL768105 EFZ12262.1 GFDF01009070 JAV05014.1 GFDL01003613 JAV31432.1 GFDL01003612 JAV31433.1 GL449813 EFN81921.1 GEZM01009896 JAV94236.1 GL888742 EGI58180.1 GL437703 EFN70103.1 APGK01035299 APGK01035300 APGK01035301 KB740923 ENN78253.1 KQ981061 KYN09979.1 KB632333 ERL92684.1 GDHF01021444 GDHF01016744 GDHF01016472 GDHF01005967 JAI30870.1 JAI35570.1 JAI35842.1 JAI46347.1 JXJN01013746 QOIP01000007 RLU20273.1 KK107238 EZA54840.1 KQ976750 KYN08707.1 GBXI01006602 JAD07690.1 GEZM01009898 JAV94235.1 KQ982701 KYQ51930.1 ADTU01020397 ADTU01020398 ADTU01020399 ADTU01020400 ADTU01020401 ADTU01020402 ADTU01020403 ADTU01020404 ADTU01020405 KQ981655 KYN38614.1 KQ976507 KYM82810.1 KQ971351 EFA06448.1 KA645935 AFP60564.1 KA649454 AFP64083.1 GAMC01019902 JAB86653.1 GAMC01019903 JAB86652.1 DS231916 EDS26132.1 KK852489 KDR22763.1 CCAG010016049 CCAG010016050 CCAG010016051 CCAG010016052 CCAG010016053 CCAG010016054 CCAG010016055 CCAG010016056 CCAG010016057 CCAG010016058 AAAB01008859 EAA07757.5 KQ434809 KZC06347.1 GBYB01002434 JAG72201.1 APCN01000584 AXCN02000323 JR048215 AEY60600.1 AJWK01024885 KQ460205 KPJ16790.1 GBXI01015169 JAC99122.1 KQ414741 KOC61984.1 GAKP01013031 JAC45921.1 GAKP01013040 JAC45912.1 GAKP01013037 JAC45915.1 GAKP01013032 JAC45920.1 ATLV01011656 KE524705 KFB36105.1 CVRI01000064 CRL05221.1 FR904289 CDQ57590.1 FR904378 CDQ61358.1 GBYX01189676 JAO83286.1 RJVU01035178 ROL47925.1 HAEH01021661 SBS12688.1 ABJB010138373 ABJB010187835 ABJB010429149 ABJB010530579 ABJB010602692 ABJB010799643 ABJB010984590 DS894052 EEC15817.1 KQ040884 KKF33439.1 CR556713 CR848823 AY934775 AAY18210.1 GBBM01001863 JAC33555.1
RSAL01000009 RVE53873.1 KQ459604 KPI92055.1 JTDY01001560 KOB73527.1 GL768105 EFZ12262.1 GFDF01009070 JAV05014.1 GFDL01003613 JAV31432.1 GFDL01003612 JAV31433.1 GL449813 EFN81921.1 GEZM01009896 JAV94236.1 GL888742 EGI58180.1 GL437703 EFN70103.1 APGK01035299 APGK01035300 APGK01035301 KB740923 ENN78253.1 KQ981061 KYN09979.1 KB632333 ERL92684.1 GDHF01021444 GDHF01016744 GDHF01016472 GDHF01005967 JAI30870.1 JAI35570.1 JAI35842.1 JAI46347.1 JXJN01013746 QOIP01000007 RLU20273.1 KK107238 EZA54840.1 KQ976750 KYN08707.1 GBXI01006602 JAD07690.1 GEZM01009898 JAV94235.1 KQ982701 KYQ51930.1 ADTU01020397 ADTU01020398 ADTU01020399 ADTU01020400 ADTU01020401 ADTU01020402 ADTU01020403 ADTU01020404 ADTU01020405 KQ981655 KYN38614.1 KQ976507 KYM82810.1 KQ971351 EFA06448.1 KA645935 AFP60564.1 KA649454 AFP64083.1 GAMC01019902 JAB86653.1 GAMC01019903 JAB86652.1 DS231916 EDS26132.1 KK852489 KDR22763.1 CCAG010016049 CCAG010016050 CCAG010016051 CCAG010016052 CCAG010016053 CCAG010016054 CCAG010016055 CCAG010016056 CCAG010016057 CCAG010016058 AAAB01008859 EAA07757.5 KQ434809 KZC06347.1 GBYB01002434 JAG72201.1 APCN01000584 AXCN02000323 JR048215 AEY60600.1 AJWK01024885 KQ460205 KPJ16790.1 GBXI01015169 JAC99122.1 KQ414741 KOC61984.1 GAKP01013031 JAC45921.1 GAKP01013040 JAC45912.1 GAKP01013037 JAC45915.1 GAKP01013032 JAC45920.1 ATLV01011656 KE524705 KFB36105.1 CVRI01000064 CRL05221.1 FR904289 CDQ57590.1 FR904378 CDQ61358.1 GBYX01189676 JAO83286.1 RJVU01035178 ROL47925.1 HAEH01021661 SBS12688.1 ABJB010138373 ABJB010187835 ABJB010429149 ABJB010530579 ABJB010602692 ABJB010799643 ABJB010984590 DS894052 EEC15817.1 KQ040884 KKF33439.1 CR556713 CR848823 AY934775 AAY18210.1 GBBM01001863 JAC33555.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000053268
UP000037510
UP000075920
+ More
UP000008237 UP000007755 UP000000311 UP000019118 UP000095300 UP000078492 UP000030742 UP000192223 UP000092443 UP000092460 UP000279307 UP000053097 UP000078542 UP000075809 UP000005205 UP000078541 UP000078540 UP000007266 UP000095301 UP000002320 UP000027135 UP000075881 UP000075901 UP000092444 UP000007062 UP000075900 UP000076502 UP000075882 UP000002358 UP000076407 UP000075840 UP000075902 UP000075886 UP000075880 UP000092461 UP000053240 UP000069272 UP000053825 UP000075903 UP000005203 UP000030765 UP000183832 UP000087266 UP000261420 UP000193380 UP000078200 UP000242638 UP000002852 UP000076408 UP000001555 UP000261380 UP000000437 UP000007635
UP000008237 UP000007755 UP000000311 UP000019118 UP000095300 UP000078492 UP000030742 UP000192223 UP000092443 UP000092460 UP000279307 UP000053097 UP000078542 UP000075809 UP000005205 UP000078541 UP000078540 UP000007266 UP000095301 UP000002320 UP000027135 UP000075881 UP000075901 UP000092444 UP000007062 UP000075900 UP000076502 UP000075882 UP000002358 UP000076407 UP000075840 UP000075902 UP000075886 UP000075880 UP000092461 UP000053240 UP000069272 UP000053825 UP000075903 UP000005203 UP000030765 UP000183832 UP000087266 UP000261420 UP000193380 UP000078200 UP000242638 UP000002852 UP000076408 UP000001555 UP000261380 UP000000437 UP000007635
PRIDE
Interpro
SUPFAM
SSF141072
SSF141072
Gene 3D
ProteinModelPortal
H9J5K5
A0A2A4K7T9
A0A2W1BTN1
A0A3S2TSG4
A0A194PLS1
A0A0L7LDH1
+ More
E9J4G5 A0A340TBD3 A0A1L8DFA0 A0A1Q3FVG2 A0A1Q3FVJ1 E2BQW7 A0A1Y1NDC9 F4X5U0 E2A904 N6TK22 A0A1I8Q2V1 A0A151ISW7 U4UFD0 A0A1W4WH66 A0A1W4W6T8 A0A1A9YLF1 A0A0K8W5L6 A0A1B0BG51 A0A3L8DJC3 A0A026WFF4 A0A195D775 A0A0A1XAD6 A0A1Y1NCB8 A0A151WVJ6 A0A158NME2 A0A151JX34 A0A151I3A1 D6WRH9 T1PCS8 T1PL58 W8AQ93 W8AL91 A0A1I8M5B8 B0WFB5 A0A067RIB9 A0A182K3A9 A0A182T391 A0A1B0GG32 Q7QCY1 A0A182RQ24 A0A154P3C3 A0A182LGC3 K7J312 A0A0C9QG73 K7J313 A0A182XHL8 A0A182HTS4 A0A182TX31 A0A182R083 V9IHT9 A0A182J9S9 A0A1B0CRK4 A0A194RHJ1 A0A182FR16 A0A0A1WL41 A0A0L7QTV0 A0A034VRI2 A0A034VUX1 A0A034VVP4 A0A034VVP9 A0A182VDX8 A0A088AB77 A0A084VDR1 A0A1J1IYD1 A0A1S3PGE9 A0A3B4VL73 A0A060VZ30 A0A1S3L856 A0A3B4VNN4 A0A3B4VJY0 A0A1A9VQY0 A0A3P9NCJ6 A0A1I8NE62 A0A3P9NCG5 A0A060W262 A0A0S7L156 A0A3N0YNZ1 A0A3B5PYK3 M4AMU3 A0A3B5RB01 A0A182XW68 A0A3B5PRY8 A0A1A8S554 B7QAE5 A0A3B5LSV5 A0A0F8CHZ0 A0A286YBI0 F6PC65 F1R4F4 Q32SG8 A0A023GL62 G3Q8Q6 G3Q8Q8 G3Q8Q4
E9J4G5 A0A340TBD3 A0A1L8DFA0 A0A1Q3FVG2 A0A1Q3FVJ1 E2BQW7 A0A1Y1NDC9 F4X5U0 E2A904 N6TK22 A0A1I8Q2V1 A0A151ISW7 U4UFD0 A0A1W4WH66 A0A1W4W6T8 A0A1A9YLF1 A0A0K8W5L6 A0A1B0BG51 A0A3L8DJC3 A0A026WFF4 A0A195D775 A0A0A1XAD6 A0A1Y1NCB8 A0A151WVJ6 A0A158NME2 A0A151JX34 A0A151I3A1 D6WRH9 T1PCS8 T1PL58 W8AQ93 W8AL91 A0A1I8M5B8 B0WFB5 A0A067RIB9 A0A182K3A9 A0A182T391 A0A1B0GG32 Q7QCY1 A0A182RQ24 A0A154P3C3 A0A182LGC3 K7J312 A0A0C9QG73 K7J313 A0A182XHL8 A0A182HTS4 A0A182TX31 A0A182R083 V9IHT9 A0A182J9S9 A0A1B0CRK4 A0A194RHJ1 A0A182FR16 A0A0A1WL41 A0A0L7QTV0 A0A034VRI2 A0A034VUX1 A0A034VVP4 A0A034VVP9 A0A182VDX8 A0A088AB77 A0A084VDR1 A0A1J1IYD1 A0A1S3PGE9 A0A3B4VL73 A0A060VZ30 A0A1S3L856 A0A3B4VNN4 A0A3B4VJY0 A0A1A9VQY0 A0A3P9NCJ6 A0A1I8NE62 A0A3P9NCG5 A0A060W262 A0A0S7L156 A0A3N0YNZ1 A0A3B5PYK3 M4AMU3 A0A3B5RB01 A0A182XW68 A0A3B5PRY8 A0A1A8S554 B7QAE5 A0A3B5LSV5 A0A0F8CHZ0 A0A286YBI0 F6PC65 F1R4F4 Q32SG8 A0A023GL62 G3Q8Q6 G3Q8Q8 G3Q8Q4
PDB
3RB7
E-value=1.12199e-13,
Score=185
Ontologies
GO
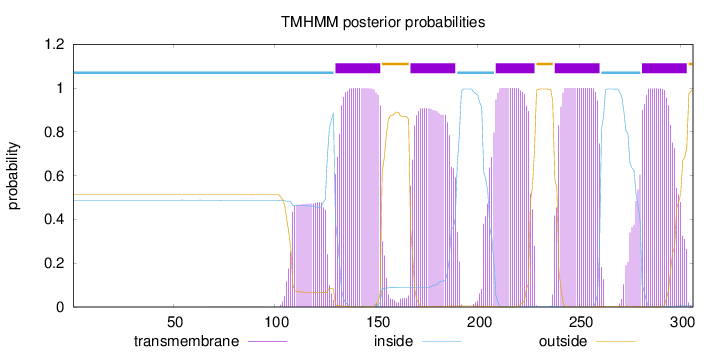
Topology
Length:
306
Number of predicted TMHs:
5
Exp number of AAs in TMHs:
116.1982
Exp number, first 60 AAs:
0.00093
Total prob of N-in:
0.48785
inside
1 - 129
TMhelix
130 - 152
outside
153 - 166
TMhelix
167 - 189
inside
190 - 208
TMhelix
209 - 228
outside
229 - 237
TMhelix
238 - 260
inside
261 - 280
TMhelix
281 - 303
outside
304 - 306
Population Genetic Test Statistics
Pi
208.973707
Theta
158.832486
Tajima's D
1.102774
CLR
0.752276
CSRT
0.693615319234038
Interpretation
Uncertain
