Gene
KWMTBOMO15008
Pre Gene Modal
BGIBMGA005515
Annotation
Snake_venom_metalloprotease_inhibitor_02A10_[Toxocara_canis]
Full name
Histone acetyltransferase
Location in the cell
Cytoplasmic Reliability : 1.498 Nuclear Reliability : 1.59
Sequence
CDS
ATGCTACAGCTGAGAATTGATTGGTTCAGTTGGTACTATTTCATAAAGTTCTTGAGAATTGATTGGTTCAAAGCAAAACAAGAAATACTACAGCTGAGAATTGATTGGTTCATTTCACAACTGAGAATCGACTGGTATGAGGAAGTTTCACAACTGAGAATCTACTGGTACGAGGAAGTTTCACAACTGAGAATCGACTGGTACGAGGAAGTTTCACAACTGAGAATCGACTGGTACGAGGAAGTTTCACAAGAATCGACTGGTACGACTGTCTCACTGAGAATCTACTGGTACGAGGAAGTTTCACAACTGAGAATCGACTGGTACGAGGAAGTTTCACAACTGAGAATCGACTGGTACGAGAAAGTCTCACAACTGAGAATCGACTGGTACGAGGAAGTTTCACAACTGAGAATCGACTGGTACGAGAAAGTCTCACAACTGAGAATCGACTGGTACGAGGAAGTTTCACAACTGAGAATCGACTGGTATGAGGAAGTTTCACAACTGAGAATCTACTGGTACGAGGAAGTTTCACAACTGAGAATCGACTGGTACGAGGAAGTTTCACAACTGAGAATCGACTGGTACGAGGAAGTTTCACAACTGAGAATCGACTGGTACGAGGAAGTTTCACAACTGAGAATCTACTGGTACGAGAAAGTCTCACAACTGAGAATCGACTGGTACGAGGAAGTTTCACAACTGAGAATCGACTGGTATGAGGAAGTTTCACAACTGAGAATCTACTGGTACGAGGAAGTTTCACAACTGAGAATCTACTGGTACGAGGAAGTTTCACAACTGAGAATCTACTGTTTCACAACTGAGAATCGACTGGCTGTGGCAGTTTCACAACTGAGAATCGACTGGTACGAGGAAGTTTCACAACTGAGAATCGACTGGTACGAGGAAGTTTCACAACTGAGAATTGACTGGCTCGAGGAAGTTTCACAACTGAGAATTGACTGGCTCGAGGAAGTTTCACAACTGAGAATTGACTGGCTCGAGGAAGTTTCACAACTGAGAATTGACTGGCTCGAGGAAGTTTCACAACTGAGAATTGACTGGCTCGAGGAAGTTTCACAACTGAGAATTGACTGGCTCGAGGAAGTTTCACAACTGAGAATTGACTGGCTCGAGGAAGTTTCACAACTGAGAATTGACTGGCTGAGGCAGTTTCACAACTGA
Protein
MLQLRIDWFSWYYFIKFLRIDWFKAKQEILQLRIDWFISQLRIDWYEEVSQLRIYWYEEVSQLRIDWYEEVSQLRIDWYEEVSQESTGTTVSLRIYWYEEVSQLRIDWYEEVSQLRIDWYEKVSQLRIDWYEEVSQLRIDWYEKVSQLRIDWYEEVSQLRIDWYEEVSQLRIYWYEEVSQLRIDWYEEVSQLRIDWYEEVSQLRIDWYEEVSQLRIYWYEKVSQLRIDWYEEVSQLRIDWYEEVSQLRIYWYEEVSQLRIYWYEEVSQLRIYCFTTENRLAVAVSQLRIDWYEEVSQLRIDWYEEVSQLRIDWLEEVSQLRIDWLEEVSQLRIDWLEEVSQLRIDWLEEVSQLRIDWLEEVSQLRIDWLEEVSQLRIDWLEEVSQLRIDWLRQFHN
Summary
Catalytic Activity
acetyl-CoA + L-lysyl-[protein] = CoA + H(+) + N(6)-acetyl-L-lysyl-[protein]
Similarity
Belongs to the apolipoprotein A1/A4/E family.
Belongs to the MYST (SAS/MOZ) family.
Belongs to the MYST (SAS/MOZ) family.
Uniprot
U3KCW3
A0A0B2VMD4
R7U0J8
S4RXQ5
S4RXQ7
G3WCV9
+ More
Q26471 A0A2H5TSX5 A0A210R2P1 L7MJH5 L7MLY6 A0A210PTE2 A0A1S4EAX3 A0A1S2WWI9 A0A0L8IBP8 A0A210QIL9 A3IXR4 A0A0Q6PWX5 A0A2M9CIN0 A0A1N6LHG3 A0A0N1J1P7 A0A0G4GLB3 N1RTP2 A0A1I8IMM3 U3P899 A7SCS1 A0A1H8EW50 A0A0Q8V013 E1SQ57 A0A1E2SI79 Q6AEU7 G4YXS5 A0A0Q5LR90 A0A1S3MQI1 A0A2M9KWI1 A7RQU1 A0A1M3M5W9
Q26471 A0A2H5TSX5 A0A210R2P1 L7MJH5 L7MLY6 A0A210PTE2 A0A1S4EAX3 A0A1S2WWI9 A0A0L8IBP8 A0A210QIL9 A3IXR4 A0A0Q6PWX5 A0A2M9CIN0 A0A1N6LHG3 A0A0N1J1P7 A0A0G4GLB3 N1RTP2 A0A1I8IMM3 U3P899 A7SCS1 A0A1H8EW50 A0A0Q8V013 E1SQ57 A0A1E2SI79 Q6AEU7 G4YXS5 A0A0Q5LR90 A0A1S3MQI1 A0A2M9KWI1 A7RQU1 A0A1M3M5W9
EC Number
2.3.1.48
Pubmed
EMBL
AGTO01002257
JPKZ01001394
KHN82205.1
AMQN01001954
KB306824
ELT99529.1
+ More
AEFK01020302 L76606 AAC37266.1 BDIQ01000146 GBC45694.1 NEDP02000713 OWF55269.1 GACK01000929 JAA64105.1 GACK01000815 JAA64219.1 NEDP02005512 OWF39725.1 JRXS01000006 OIN83919.1 KQ416137 KOF98450.1 NEDP02003494 OWF48539.1 AAXW01000069 EAZ88717.1 LMCU01000001 KQV07727.1 PGFF01000001 PJJ71796.1 FSRV01000002 SIO68116.1 JPDT01000262 KPA18932.1 CDMZ01001326 CEM30873.1 KB726545 EMT68731.1 CP006734 AGW41162.1 DS469625 EDO38449.1 FOCN01000005 SEN22958.1 LMIO01000002 KRC50843.1 CP002209 ADN76829.1 LNZG01000047 ODA89384.1 AE016822 AAT89098.1 JH159152 EGZ25068.1 LMOZ01000002 KQR52795.1 NNBO01000010 PJN23480.1 DS469529 EDO46165.1 MKVJ01000001 OJX81530.1
AEFK01020302 L76606 AAC37266.1 BDIQ01000146 GBC45694.1 NEDP02000713 OWF55269.1 GACK01000929 JAA64105.1 GACK01000815 JAA64219.1 NEDP02005512 OWF39725.1 JRXS01000006 OIN83919.1 KQ416137 KOF98450.1 NEDP02003494 OWF48539.1 AAXW01000069 EAZ88717.1 LMCU01000001 KQV07727.1 PGFF01000001 PJJ71796.1 FSRV01000002 SIO68116.1 JPDT01000262 KPA18932.1 CDMZ01001326 CEM30873.1 KB726545 EMT68731.1 CP006734 AGW41162.1 DS469625 EDO38449.1 FOCN01000005 SEN22958.1 LMIO01000002 KRC50843.1 CP002209 ADN76829.1 LNZG01000047 ODA89384.1 AE016822 AAT89098.1 JH159152 EGZ25068.1 LMOZ01000002 KQR52795.1 NNBO01000010 PJN23480.1 DS469529 EDO46165.1 MKVJ01000001 OJX81530.1
Proteomes
UP000016665
UP000031036
UP000014760
UP000245300
UP000007648
UP000236242
+ More
UP000242188 UP000079169 UP000181723 UP000053454 UP000003781 UP000051914 UP000228758 UP000184700 UP000037988 UP000016929 UP000095280 UP000016743 UP000001593 UP000198890 UP000051819 UP000006683 UP000094426 UP000001306 UP000002640 UP000050966 UP000087266 UP000231957 UP000184281
UP000242188 UP000079169 UP000181723 UP000053454 UP000003781 UP000051914 UP000228758 UP000184700 UP000037988 UP000016929 UP000095280 UP000016743 UP000001593 UP000198890 UP000051819 UP000006683 UP000094426 UP000001306 UP000002640 UP000050966 UP000087266 UP000231957 UP000184281
Pfam
Interpro
IPR000074
ApoA_E
+ More
IPR004155 PBS_lyase_HEAT
IPR009818 Ataxin-2_C
IPR036388 WH-like_DNA-bd_sf
IPR040706 Zf-MYST
IPR011011 Znf_FYVE_PHD
IPR005818 Histone_H1/H5_H15
IPR036390 WH_DNA-bd_sf
IPR002717 HAT_MYST-type
IPR001965 Znf_PHD
IPR013083 Znf_RING/FYVE/PHD
IPR019787 Znf_PHD-finger
IPR016181 Acyl_CoA_acyltransferase
IPR036322 WD40_repeat_dom_sf
IPR032675 LRR_dom_sf
IPR013783 Ig-like_fold
IPR007110 Ig-like_dom
IPR003599 Ig_sub
IPR013098 Ig_I-set
IPR036179 Ig-like_dom_sf
IPR003598 Ig_sub2
IPR003409 MORN
IPR016024 ARM-type_fold
IPR019933 DivIVA_domain
IPR000408 Reg_chr_condens
IPR009091 RCC1/BLIP-II
IPR004155 PBS_lyase_HEAT
IPR009818 Ataxin-2_C
IPR036388 WH-like_DNA-bd_sf
IPR040706 Zf-MYST
IPR011011 Znf_FYVE_PHD
IPR005818 Histone_H1/H5_H15
IPR036390 WH_DNA-bd_sf
IPR002717 HAT_MYST-type
IPR001965 Znf_PHD
IPR013083 Znf_RING/FYVE/PHD
IPR019787 Znf_PHD-finger
IPR016181 Acyl_CoA_acyltransferase
IPR036322 WD40_repeat_dom_sf
IPR032675 LRR_dom_sf
IPR013783 Ig-like_fold
IPR007110 Ig-like_dom
IPR003599 Ig_sub
IPR013098 Ig_I-set
IPR036179 Ig-like_dom_sf
IPR003598 Ig_sub2
IPR003409 MORN
IPR016024 ARM-type_fold
IPR019933 DivIVA_domain
IPR000408 Reg_chr_condens
IPR009091 RCC1/BLIP-II
SUPFAM
ProteinModelPortal
U3KCW3
A0A0B2VMD4
R7U0J8
S4RXQ5
S4RXQ7
G3WCV9
+ More
Q26471 A0A2H5TSX5 A0A210R2P1 L7MJH5 L7MLY6 A0A210PTE2 A0A1S4EAX3 A0A1S2WWI9 A0A0L8IBP8 A0A210QIL9 A3IXR4 A0A0Q6PWX5 A0A2M9CIN0 A0A1N6LHG3 A0A0N1J1P7 A0A0G4GLB3 N1RTP2 A0A1I8IMM3 U3P899 A7SCS1 A0A1H8EW50 A0A0Q8V013 E1SQ57 A0A1E2SI79 Q6AEU7 G4YXS5 A0A0Q5LR90 A0A1S3MQI1 A0A2M9KWI1 A7RQU1 A0A1M3M5W9
Q26471 A0A2H5TSX5 A0A210R2P1 L7MJH5 L7MLY6 A0A210PTE2 A0A1S4EAX3 A0A1S2WWI9 A0A0L8IBP8 A0A210QIL9 A3IXR4 A0A0Q6PWX5 A0A2M9CIN0 A0A1N6LHG3 A0A0N1J1P7 A0A0G4GLB3 N1RTP2 A0A1I8IMM3 U3P899 A7SCS1 A0A1H8EW50 A0A0Q8V013 E1SQ57 A0A1E2SI79 Q6AEU7 G4YXS5 A0A0Q5LR90 A0A1S3MQI1 A0A2M9KWI1 A7RQU1 A0A1M3M5W9
Ontologies
KEGG
GO
Topology
Subcellular location
Secreted
Nucleus
Nucleus
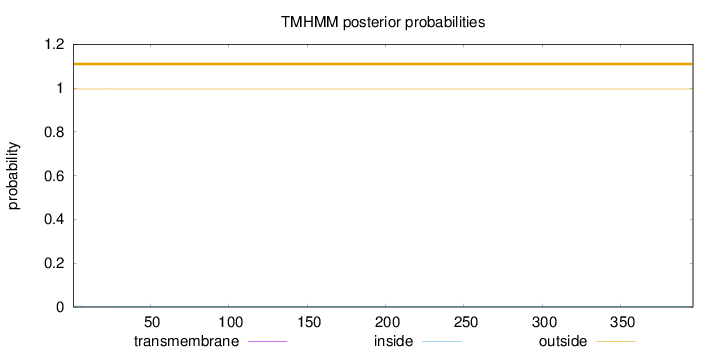
Length:
396
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00594
Exp number, first 60 AAs:
0.00594
Total prob of N-in:
0.00412
outside
1 - 396
Population Genetic Test Statistics
Pi
315.110238
Theta
213.797788
Tajima's D
1.786749
CLR
0
CSRT
0.848057597120144
Interpretation
Uncertain
