Gene
KWMTBOMO15005
Pre Gene Modal
BGIBMGA004794
Annotation
PREDICTED:_sodium/calcium_exchanger_2_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 1.477
Sequence
CDS
ATGCGGCCCGCGCTCGCCGCGATCGCGGCCTGCTGCGTGGCGGCACGCGCCGAGCCCCCCTCGCGCCTCGCACCCCTCAACACCACCCCCCAGCACGTTCTCCAGTGCTCCGATGGCCTCGTGCTGCCCGCCTGGAGGCCTCTCGAAGATGTCCCCACTTACCAAATCGCGATACGCGGCTTCGTCTACTTTCTTGCGCTCATGTACCTCTTCATTGGGGTGTCTATTGTCTCTGATCGGTTTATGGCCGCCATAGAAGTTATCACGTCCAGAGAAAAAGAGGTGCGCGTGCGGCGCAACGGCAGTGAACAGATCATAGTTGTTCGAGTCTGGAATGAGACCGTTGCCAATTTGACGTTGATGGCTCTCGGATCGAGTGCACCTGAGATACTCTTATCAGTAATCGAGATAGCCGGAAAGAATTTCGAAGCCGGAGATTTGGGCCCCGGCACCATCGTGGGTTCTGCTGCCTATAATCTTTTTGTCATAATCGCCATCTGCGTTGTGGTGATCCCGGATGGAGAAGTGCGCAAGATAAAACATCTTCGAGTATTCTTAGTGACCGCGACTTGGTCGATATTCGCCTATGTATGGCTATATATGATTTTGGCGGTTATATCACCAAGTGAAGTCGAAGTATGGGAGGGTGCGGTCACTTTCTTGTTCTTCCCGGCGACTGTCATGACAGCTTATATTGCTGATCGTCGCCTGCTCGTCTATAAATATCTCAAGAAAGGCTACAGAGTGAACGAGAGAGGCGTCATAGTTGAAGCCGAGGGTGACGGTTCTCTAGAGATGGCGCTGAAGCCTGGCGGAGACGATTTTATGTCTGATGACGACGAAGGCCGAGAACTAGAGAAGTCACGGAAAGAATACGTATCAGTTCTCCGCGAACTGAGAAGACAGCATCCAGACGAAGATTTAGAAACCGTTGAGAGAATGGCTTTGGAAACGTTAATGGCAAGTGGTCCTAAATCACGAGCGTTCTACAGAGTTGCGGCGACTCGTAAAATGACCGGTGGCGGTGATTTTTCGCGGAAAATCTCCGAGAGGGCCCAGAGCGATTTGAGCGAGGTAAAAGCGGAGTTACAGAGACGAGATTCTGGTTCGGCGTCAATTGAGCCGCGCGGTCCAAACGTTCGTTTCGATCCTGACCACTACACCGTCATGGAGAACTGCGGCTCGTTCGAAGTTCGAGTTGTAAGGGCTGGAGATCTTAACGGTGACGTCACTGTTCAGTACACCACCGAAGATGGGACCGCTGAAGCTGGCTCAGATTATGTGACCGCTCAAGGATTATTGCATTTCAAACACGGAGAAACGGAGAAGACGTTTAGAGTTCAAGTCATTGATGATGATGTTTTTGAGGAAGATGAGCATTTTTACGTTCGTCTGTCATCGCCAAAGGGCGCTTCCTTGGCTTCTCCGTCTACCGCTACTGTAGTGATACTCGACGATGACCACTCCGGCGTATTTTCGTTCCCTCAACGCGATTTCGAGCTGACAGAGTCCGTTGGGACATATGATCTGAAGGTGGTGAGGACAGCTGGCGCCCGCGGGAAGGTCAGGATTCCTTACTGGACCGAGGACGGCACAGCTAAGGGAGGCGCTCAGTACGAGATTGCACAAGGCCACATCTTCTTCGAAAACAACGAGACCGAGTAA
Protein
MRPALAAIAACCVAARAEPPSRLAPLNTTPQHVLQCSDGLVLPAWRPLEDVPTYQIAIRGFVYFLALMYLFIGVSIVSDRFMAAIEVITSREKEVRVRRNGSEQIIVVRVWNETVANLTLMALGSSAPEILLSVIEIAGKNFEAGDLGPGTIVGSAAYNLFVIIAICVVVIPDGEVRKIKHLRVFLVTATWSIFAYVWLYMILAVISPSEVEVWEGAVTFLFFPATVMTAYIADRRLLVYKYLKKGYRVNERGVIVEAEGDGSLEMALKPGGDDFMSDDDEGRELEKSRKEYVSVLRELRRQHPDEDLETVERMALETLMASGPKSRAFYRVAATRKMTGGGDFSRKISERAQSDLSEVKAELQRRDSGSASIEPRGPNVRFDPDHYTVMENCGSFEVRVVRAGDLNGDVTVQYTTEDGTAEAGSDYVTAQGLLHFKHGETEKTFRVQVIDDDVFEEDEHFYVRLSSPKGASLASPSTATVVILDDDHSGVFSFPQRDFELTESVGTYDLKVVRTAGARGKVRIPYWTEDGTAKGGAQYEIAQGHIFFENNETE
Summary
Similarity
Belongs to the Ca(2+):cation antiporter (CaCA) (TC 2.A.19) family.
Uniprot
H9J5K4
A0A0L7L0T9
A0A2W1BVS7
A0A2A4K8J9
A0A2H1VYJ5
A0A194PLS1
+ More
A0A194RHJ1 A0A182JJZ5 A0A182FR14 A0A084VDP7 A0A2M4D0D9 W5JPW4 A0A182LUI7 A0A1W4W6T8 A0A1W4WH66 A0A182LGD6 A0A182PSN5 A0A182XHL9 A0A182N736 A0A1Q3FVJ1 A0A182TGL3 A0A182HTS1 A0A182RQ22 A0A1Y1NCB8 A0A1Q3FVG2 A0A182VKI1 B0WFC7 T1PL58 A0A1I8M5B8 A0A182XW65 T1PCS8 A0A1Y1NF08 A0A182QRM5 Q7QCY1 A0A182SRE0 A0A340TBD3 A0A1I8NM23 A0A182GWS3 A0A0L0C7G8 A0A088AB77 Q16I58 A0A1A9VQX7 A0A336MKN7 A0A336KYC1 A0A2P8YJ54 A0A1B0GG32 A0A1B0BG46 A0A1A9YLF2 A0A1A9ZK36 D6WRH9 A0A1A9WFS1 A0A034VVP4 A0A034VUX1 A0A067RIB9 A0A034VRI2 K7J313 K7J312 A0A034VVP9 A0A0K8V717 A0A0K8V0D8 A0A0A1WL41 E2BQW9 A0A1B0F0A8 A0A0C9QG73 A0A154P3C3 A0A0L7QTV0 A0A151JX34 E2A905 A0A195D775 A0A026WFF4 A0A3L8DJC3 A0A151J7Z1 A0A158NME2 E9J4G4 A0A151I3A1 W8AQ93 W8AL91 A0A1B0CQJ3 A0A1J1IYH5 A0A151WVJ6 N6TD33 E9GI59 F4X5T9 A0A0Q9WT62 A0A3B0K369 B4NBR4 A0A3B0K4Y3 A0A0Q9X3H3 B4R1P3 Q24413 A0A0B4LHD7 Q9VDG5 A0A0B4K6R7 A0A0B4K6E2 A0A0B4K6A6 A0A0B4K790 A0A0R1E453 A0A0R1E1M0 B4PQ94 A0A1W4VYS2
A0A194RHJ1 A0A182JJZ5 A0A182FR14 A0A084VDP7 A0A2M4D0D9 W5JPW4 A0A182LUI7 A0A1W4W6T8 A0A1W4WH66 A0A182LGD6 A0A182PSN5 A0A182XHL9 A0A182N736 A0A1Q3FVJ1 A0A182TGL3 A0A182HTS1 A0A182RQ22 A0A1Y1NCB8 A0A1Q3FVG2 A0A182VKI1 B0WFC7 T1PL58 A0A1I8M5B8 A0A182XW65 T1PCS8 A0A1Y1NF08 A0A182QRM5 Q7QCY1 A0A182SRE0 A0A340TBD3 A0A1I8NM23 A0A182GWS3 A0A0L0C7G8 A0A088AB77 Q16I58 A0A1A9VQX7 A0A336MKN7 A0A336KYC1 A0A2P8YJ54 A0A1B0GG32 A0A1B0BG46 A0A1A9YLF2 A0A1A9ZK36 D6WRH9 A0A1A9WFS1 A0A034VVP4 A0A034VUX1 A0A067RIB9 A0A034VRI2 K7J313 K7J312 A0A034VVP9 A0A0K8V717 A0A0K8V0D8 A0A0A1WL41 E2BQW9 A0A1B0F0A8 A0A0C9QG73 A0A154P3C3 A0A0L7QTV0 A0A151JX34 E2A905 A0A195D775 A0A026WFF4 A0A3L8DJC3 A0A151J7Z1 A0A158NME2 E9J4G4 A0A151I3A1 W8AQ93 W8AL91 A0A1B0CQJ3 A0A1J1IYH5 A0A151WVJ6 N6TD33 E9GI59 F4X5T9 A0A0Q9WT62 A0A3B0K369 B4NBR4 A0A3B0K4Y3 A0A0Q9X3H3 B4R1P3 Q24413 A0A0B4LHD7 Q9VDG5 A0A0B4K6R7 A0A0B4K6E2 A0A0B4K6A6 A0A0B4K790 A0A0R1E453 A0A0R1E1M0 B4PQ94 A0A1W4VYS2
Pubmed
19121390
26227816
28756777
26354079
24438588
20920257
+ More
23761445 20966253 28004739 25315136 25244985 12364791 14747013 17210077 26483478 26108605 17510324 29403074 18362917 19820115 25348373 24845553 20075255 25830018 20798317 24508170 30249741 21347285 21282665 24495485 23537049 21292972 21719571 17994087 8659862 9252464 19361442 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 22000518 26109357 26109356 17550304
23761445 20966253 28004739 25315136 25244985 12364791 14747013 17210077 26483478 26108605 17510324 29403074 18362917 19820115 25348373 24845553 20075255 25830018 20798317 24508170 30249741 21347285 21282665 24495485 23537049 21292972 21719571 17994087 8659862 9252464 19361442 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 22000518 26109357 26109356 17550304
EMBL
BABH01025184
JTDY01003719
KOB69097.1
KZ149903
PZC78441.1
NWSH01000067
+ More
PCG80000.1 ODYU01005227 SOQ45878.1 KQ459604 KPI92055.1 KQ460205 KPJ16790.1 ATLV01011641 ATLV01011642 ATLV01011643 ATLV01011644 ATLV01011645 ATLV01011646 ATLV01011647 ATLV01011648 KE524705 KFB36091.1 GGFL01006763 MBW70941.1 ADMH02000448 ETN66432.1 AXCM01011352 GFDL01003612 JAV31433.1 APCN01000583 GEZM01009898 JAV94235.1 GFDL01003613 JAV31432.1 DS231916 EDS26144.1 KA649454 AFP64083.1 KA645935 AFP60564.1 GEZM01009896 JAV94237.1 AXCN02000324 AAAB01008859 EAA07757.5 JXUM01094026 KQ564097 KXJ72802.1 JRES01000809 KNC28220.1 CH478105 EAT33944.1 UFQS01001222 UFQT01001222 SSX09826.1 SSX29549.1 SSX09827.1 SSX29550.1 PYGN01000557 PSN44272.1 CCAG010016049 CCAG010016050 CCAG010016051 CCAG010016052 CCAG010016053 CCAG010016054 CCAG010016055 CCAG010016056 CCAG010016057 CCAG010016058 JXJN01013744 JXJN01026459 KQ971351 EFA06448.1 GAKP01013037 JAC45915.1 GAKP01013040 JAC45912.1 KK852489 KDR22763.1 GAKP01013031 JAC45921.1 GAKP01013032 JAC45920.1 GDHF01026480 GDHF01025859 GDHF01017628 GDHF01003041 JAI25834.1 JAI26455.1 JAI34686.1 JAI49273.1 GDHF01020071 JAI32243.1 GBXI01015169 JAC99122.1 GL449813 EFN81923.1 AJVK01017886 GBYB01002434 JAG72201.1 KQ434809 KZC06347.1 KQ414741 KOC61984.1 KQ981655 KYN38614.1 GL437703 EFN70104.1 KQ976750 KYN08707.1 KK107238 EZA54840.1 QOIP01000007 RLU20273.1 KQ979602 KYN20513.1 ADTU01020397 ADTU01020398 ADTU01020399 ADTU01020400 ADTU01020401 ADTU01020402 ADTU01020403 ADTU01020404 ADTU01020405 GL768105 EFZ12237.1 KQ976507 KYM82810.1 GAMC01019902 JAB86653.1 GAMC01019903 JAB86652.1 AJWK01023677 CVRI01000064 CRL05207.1 KQ982701 KYQ51930.1 APGK01035308 KB740923 KB632333 ENN78254.1 ERL92685.1 GL732546 EFX80619.1 GL888742 EGI58179.1 CH964232 KRF99413.1 OUUW01000013 SPP87763.1 EDW81228.1 SPP87762.1 KRF99414.1 CM000364 EDX12152.1 L39835 AAB50166.1 AE014297 AHN57433.1 AY118680 BT011420 AAF55829.1 AAM50540.1 AAN13844.1 AAN13845.1 AAR96212.1 AFH06530.1 AFH06533.1 AFH06532.1 AFH06531.1 CM000160 KRK02992.1 KRK02995.1 KRK02991.1 EDW96203.2 KRK02989.1 KRK02993.1 KRK02994.1 KRK02996.1
PCG80000.1 ODYU01005227 SOQ45878.1 KQ459604 KPI92055.1 KQ460205 KPJ16790.1 ATLV01011641 ATLV01011642 ATLV01011643 ATLV01011644 ATLV01011645 ATLV01011646 ATLV01011647 ATLV01011648 KE524705 KFB36091.1 GGFL01006763 MBW70941.1 ADMH02000448 ETN66432.1 AXCM01011352 GFDL01003612 JAV31433.1 APCN01000583 GEZM01009898 JAV94235.1 GFDL01003613 JAV31432.1 DS231916 EDS26144.1 KA649454 AFP64083.1 KA645935 AFP60564.1 GEZM01009896 JAV94237.1 AXCN02000324 AAAB01008859 EAA07757.5 JXUM01094026 KQ564097 KXJ72802.1 JRES01000809 KNC28220.1 CH478105 EAT33944.1 UFQS01001222 UFQT01001222 SSX09826.1 SSX29549.1 SSX09827.1 SSX29550.1 PYGN01000557 PSN44272.1 CCAG010016049 CCAG010016050 CCAG010016051 CCAG010016052 CCAG010016053 CCAG010016054 CCAG010016055 CCAG010016056 CCAG010016057 CCAG010016058 JXJN01013744 JXJN01026459 KQ971351 EFA06448.1 GAKP01013037 JAC45915.1 GAKP01013040 JAC45912.1 KK852489 KDR22763.1 GAKP01013031 JAC45921.1 GAKP01013032 JAC45920.1 GDHF01026480 GDHF01025859 GDHF01017628 GDHF01003041 JAI25834.1 JAI26455.1 JAI34686.1 JAI49273.1 GDHF01020071 JAI32243.1 GBXI01015169 JAC99122.1 GL449813 EFN81923.1 AJVK01017886 GBYB01002434 JAG72201.1 KQ434809 KZC06347.1 KQ414741 KOC61984.1 KQ981655 KYN38614.1 GL437703 EFN70104.1 KQ976750 KYN08707.1 KK107238 EZA54840.1 QOIP01000007 RLU20273.1 KQ979602 KYN20513.1 ADTU01020397 ADTU01020398 ADTU01020399 ADTU01020400 ADTU01020401 ADTU01020402 ADTU01020403 ADTU01020404 ADTU01020405 GL768105 EFZ12237.1 KQ976507 KYM82810.1 GAMC01019902 JAB86653.1 GAMC01019903 JAB86652.1 AJWK01023677 CVRI01000064 CRL05207.1 KQ982701 KYQ51930.1 APGK01035308 KB740923 KB632333 ENN78254.1 ERL92685.1 GL732546 EFX80619.1 GL888742 EGI58179.1 CH964232 KRF99413.1 OUUW01000013 SPP87763.1 EDW81228.1 SPP87762.1 KRF99414.1 CM000364 EDX12152.1 L39835 AAB50166.1 AE014297 AHN57433.1 AY118680 BT011420 AAF55829.1 AAM50540.1 AAN13844.1 AAN13845.1 AAR96212.1 AFH06530.1 AFH06533.1 AFH06532.1 AFH06531.1 CM000160 KRK02992.1 KRK02995.1 KRK02991.1 EDW96203.2 KRK02989.1 KRK02993.1 KRK02994.1 KRK02996.1
Proteomes
UP000005204
UP000037510
UP000218220
UP000053268
UP000053240
UP000075880
+ More
UP000069272 UP000030765 UP000000673 UP000075883 UP000192223 UP000075882 UP000075885 UP000076407 UP000075884 UP000075902 UP000075840 UP000075900 UP000075903 UP000002320 UP000095301 UP000076408 UP000075886 UP000007062 UP000075901 UP000075920 UP000095300 UP000069940 UP000249989 UP000037069 UP000005203 UP000008820 UP000078200 UP000245037 UP000092444 UP000092460 UP000092443 UP000092445 UP000007266 UP000091820 UP000027135 UP000002358 UP000008237 UP000092462 UP000076502 UP000053825 UP000078541 UP000000311 UP000078542 UP000053097 UP000279307 UP000078492 UP000005205 UP000078540 UP000092461 UP000183832 UP000075809 UP000019118 UP000030742 UP000000305 UP000007755 UP000007798 UP000268350 UP000000304 UP000000803 UP000002282 UP000192221
UP000069272 UP000030765 UP000000673 UP000075883 UP000192223 UP000075882 UP000075885 UP000076407 UP000075884 UP000075902 UP000075840 UP000075900 UP000075903 UP000002320 UP000095301 UP000076408 UP000075886 UP000007062 UP000075901 UP000075920 UP000095300 UP000069940 UP000249989 UP000037069 UP000005203 UP000008820 UP000078200 UP000245037 UP000092444 UP000092460 UP000092443 UP000092445 UP000007266 UP000091820 UP000027135 UP000002358 UP000008237 UP000092462 UP000076502 UP000053825 UP000078541 UP000000311 UP000078542 UP000053097 UP000279307 UP000078492 UP000005205 UP000078540 UP000092461 UP000183832 UP000075809 UP000019118 UP000030742 UP000000305 UP000007755 UP000007798 UP000268350 UP000000304 UP000000803 UP000002282 UP000192221
Interpro
SUPFAM
SSF141072
SSF141072
Gene 3D
ProteinModelPortal
H9J5K4
A0A0L7L0T9
A0A2W1BVS7
A0A2A4K8J9
A0A2H1VYJ5
A0A194PLS1
+ More
A0A194RHJ1 A0A182JJZ5 A0A182FR14 A0A084VDP7 A0A2M4D0D9 W5JPW4 A0A182LUI7 A0A1W4W6T8 A0A1W4WH66 A0A182LGD6 A0A182PSN5 A0A182XHL9 A0A182N736 A0A1Q3FVJ1 A0A182TGL3 A0A182HTS1 A0A182RQ22 A0A1Y1NCB8 A0A1Q3FVG2 A0A182VKI1 B0WFC7 T1PL58 A0A1I8M5B8 A0A182XW65 T1PCS8 A0A1Y1NF08 A0A182QRM5 Q7QCY1 A0A182SRE0 A0A340TBD3 A0A1I8NM23 A0A182GWS3 A0A0L0C7G8 A0A088AB77 Q16I58 A0A1A9VQX7 A0A336MKN7 A0A336KYC1 A0A2P8YJ54 A0A1B0GG32 A0A1B0BG46 A0A1A9YLF2 A0A1A9ZK36 D6WRH9 A0A1A9WFS1 A0A034VVP4 A0A034VUX1 A0A067RIB9 A0A034VRI2 K7J313 K7J312 A0A034VVP9 A0A0K8V717 A0A0K8V0D8 A0A0A1WL41 E2BQW9 A0A1B0F0A8 A0A0C9QG73 A0A154P3C3 A0A0L7QTV0 A0A151JX34 E2A905 A0A195D775 A0A026WFF4 A0A3L8DJC3 A0A151J7Z1 A0A158NME2 E9J4G4 A0A151I3A1 W8AQ93 W8AL91 A0A1B0CQJ3 A0A1J1IYH5 A0A151WVJ6 N6TD33 E9GI59 F4X5T9 A0A0Q9WT62 A0A3B0K369 B4NBR4 A0A3B0K4Y3 A0A0Q9X3H3 B4R1P3 Q24413 A0A0B4LHD7 Q9VDG5 A0A0B4K6R7 A0A0B4K6E2 A0A0B4K6A6 A0A0B4K790 A0A0R1E453 A0A0R1E1M0 B4PQ94 A0A1W4VYS2
A0A194RHJ1 A0A182JJZ5 A0A182FR14 A0A084VDP7 A0A2M4D0D9 W5JPW4 A0A182LUI7 A0A1W4W6T8 A0A1W4WH66 A0A182LGD6 A0A182PSN5 A0A182XHL9 A0A182N736 A0A1Q3FVJ1 A0A182TGL3 A0A182HTS1 A0A182RQ22 A0A1Y1NCB8 A0A1Q3FVG2 A0A182VKI1 B0WFC7 T1PL58 A0A1I8M5B8 A0A182XW65 T1PCS8 A0A1Y1NF08 A0A182QRM5 Q7QCY1 A0A182SRE0 A0A340TBD3 A0A1I8NM23 A0A182GWS3 A0A0L0C7G8 A0A088AB77 Q16I58 A0A1A9VQX7 A0A336MKN7 A0A336KYC1 A0A2P8YJ54 A0A1B0GG32 A0A1B0BG46 A0A1A9YLF2 A0A1A9ZK36 D6WRH9 A0A1A9WFS1 A0A034VVP4 A0A034VUX1 A0A067RIB9 A0A034VRI2 K7J313 K7J312 A0A034VVP9 A0A0K8V717 A0A0K8V0D8 A0A0A1WL41 E2BQW9 A0A1B0F0A8 A0A0C9QG73 A0A154P3C3 A0A0L7QTV0 A0A151JX34 E2A905 A0A195D775 A0A026WFF4 A0A3L8DJC3 A0A151J7Z1 A0A158NME2 E9J4G4 A0A151I3A1 W8AQ93 W8AL91 A0A1B0CQJ3 A0A1J1IYH5 A0A151WVJ6 N6TD33 E9GI59 F4X5T9 A0A0Q9WT62 A0A3B0K369 B4NBR4 A0A3B0K4Y3 A0A0Q9X3H3 B4R1P3 Q24413 A0A0B4LHD7 Q9VDG5 A0A0B4K6R7 A0A0B4K6E2 A0A0B4K6A6 A0A0B4K790 A0A0R1E453 A0A0R1E1M0 B4PQ94 A0A1W4VYS2
PDB
3RB7
E-value=1.34009e-54,
Score=540
Ontologies
GO
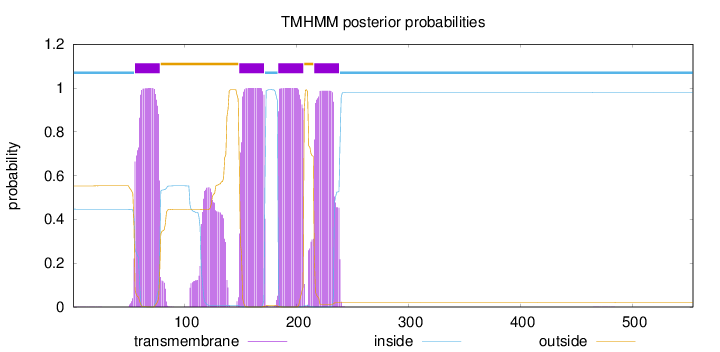
Topology
SignalP
Position: 1 - 17,
Likelihood: 0.951447
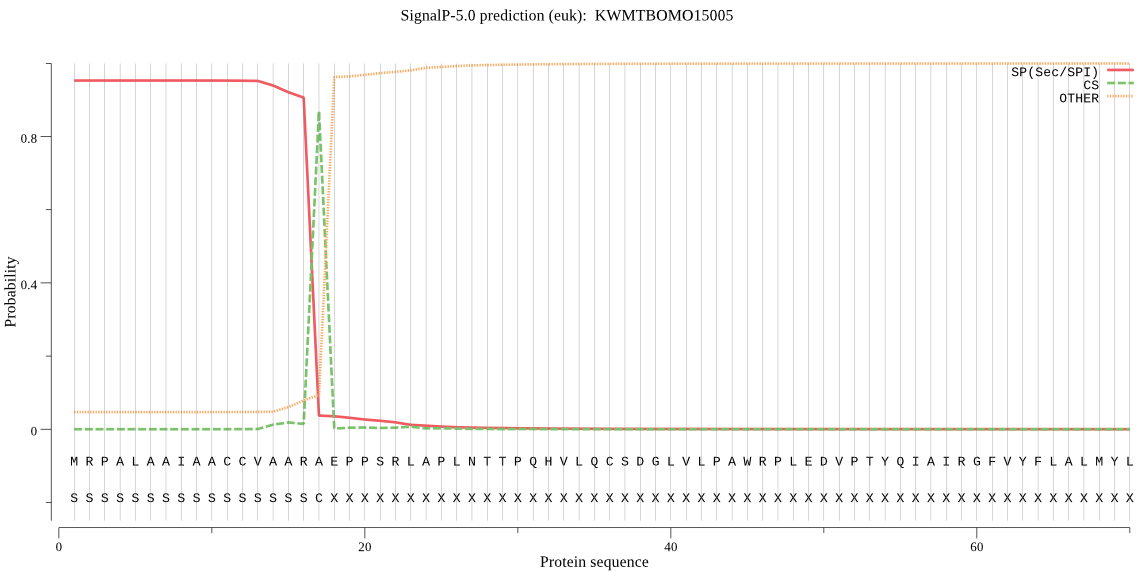
Length:
554
Number of predicted TMHs:
4
Exp number of AAs in TMHs:
100.2029
Exp number, first 60 AAs:
3.99661
Total prob of N-in:
0.44712
inside
1 - 55
TMhelix
56 - 78
outside
79 - 148
TMhelix
149 - 171
inside
172 - 183
TMhelix
184 - 206
outside
207 - 215
TMhelix
216 - 238
inside
239 - 554
Population Genetic Test Statistics
Pi
263.212695
Theta
236.149198
Tajima's D
0.28211
CLR
0
CSRT
0.451977401129944
Interpretation
Uncertain
