Gene
KWMTBOMO15000
Pre Gene Modal
BGIBMGA004790
Annotation
PREDICTED:_cyclin-dependent_kinase-like_1_[Bombyx_mori]
Full name
Cyclin-dependent kinase-like 1
Alternative Name
Protein kinase p42 KKIALRE
Serine/threonine-protein kinase KKIALRE
Serine/threonine-protein kinase KKIALRE
Location in the cell
Mitochondrial Reliability : 1.526 Nuclear Reliability : 1.915
Sequence
CDS
ATGAAGAATTCAGTGAACGAGGACACCTCGAAATTCAAATTGTGGAACAACGTGGCGTCCTGCATGGAGAAATGGTTCGGTGATAAACGGCTCTGGTTATTTGGCAACAACCTGCCGCTGTTACCGAGAAGGTCACGCGCCAGTAGCCGCGCAATGGAACGCTACGAGAAACTTGCCAAGCTAGGCGAGGGCTCCTACGGTCTGGTCTACAAGTGCCGCAACAGAGACACCGGCGAGATAGTCGCGGTGAAGAAGTTCGTTGAAAACGAAGACGATCCACTCATCAGGAAGATCGCTTTGCGGGAGATACGGATGTTGAAGAACTTGAAGCATCCGAACCTTGTTAATTTAATTGAAGTTTTTCGACGGAAGCGCAAACTGCATTTGGTGTTTGAATACTGTGACCACACGGTGCTGCACGAATTGGAAAAGTATCCAGCCGGCTGCCCCGAACTACTTTCAAAGCAGATAATCTGGCAGACTTTGCAAGGGGTCGCGTACTGCCATCGGCATAATTGTATCCACCGTGACGTCAAACCAGAGAACATACTCCTCACGAGCGACGGAGTCGTCAAGCTCTGTGACTTTGGATTCGCAAGGATGATCAGTGCGTATAAAAAATTATTTCAATTACTTAAGGGTTAA
Protein
MKNSVNEDTSKFKLWNNVASCMEKWFGDKRLWLFGNNLPLLPRRSRASSRAMERYEKLAKLGEGSYGLVYKCRNRDTGEIVAVKKFVENEDDPLIRKIALREIRMLKNLKHPNLVNLIEVFRRKRKLHLVFEYCDHTVLHELEKYPAGCPELLSKQIIWQTLQGVAYCHRHNCIHRDVKPENILLTSDGVVKLCDFGFARMISAYKKLFQLLKG
Summary
Catalytic Activity
ATP + L-seryl-[protein] = ADP + H(+) + O-phospho-L-seryl-[protein]
ATP + L-threonyl-[protein] = ADP + H(+) + O-phospho-L-threonyl-[protein]
ATP + L-threonyl-[protein] = ADP + H(+) + O-phospho-L-threonyl-[protein]
Similarity
Belongs to the protein kinase superfamily.
Belongs to the protein kinase superfamily. CMGC Ser/Thr protein kinase family. CDC2/CDKX subfamily.
Belongs to the protein kinase superfamily. CMGC Ser/Thr protein kinase family. CDC2/CDKX subfamily.
Keywords
3D-structure
Alternative splicing
ATP-binding
Complete proteome
Cytoplasm
Kinase
Nucleotide-binding
Nucleus
Polymorphism
Reference proteome
Transferase
Feature
chain Cyclin-dependent kinase-like 1
splice variant In isoform 2.
sequence variant In dbSNP:rs11570814.
splice variant In isoform 2.
sequence variant In dbSNP:rs11570814.
Uniprot
A0A2H1WWL6
A0A2A4JUQ8
A0A3S2LHD6
A0A194PH83
A0A0L7LID4
H9J5K0
+ More
A0A212FPW4 D6WSF5 A0A158NAV6 A0A232EVU3 A0A0K8TJ02 A0A067R7U0 A0A2P8XRW4 A0A026WUU7 A0A2H8TLW9 E2AT42 A0A2S2NVL7 A0A1S4F8D7 A0A1B6CAC5 J9JXR5 E0VSK6 X2JD86 A0A0J9QWB6 A0A0P5K232 A0A0P5A5F0 A0A0P5CLC3 A0A182QK93 A0A0R3NZS1 A0A0P5JNY3 A0A1W4VBS3 A0A0P6AH56 A0A0P6GUD6 A0A2A3EJD4 A0A0N8EN14 A0A3L8D8P0 A0A182R375 A0A182XE57 U4U3Z9 A0A1Y1LJJ0 A0A1D2NE10 K7J404 N6TBM8 A0A2R7WIC0 A0A0K8TIH2 A0A310SXL0 A0A154P6C2 V9LIN5 F4X245 A0A195E4V6 A0A151I5W7 A0A0L7QK80 A0A195EVD1 A0A195CK25 E9G338 A0A1A9USN7 E2BPF6 A0A088ABR9 A0A0M9A4V5 A0A034V2J7 A0A2P6L9K2 A0A087UTB1 A0A226DFG1 A0A182H5B4 A0A1B0G098 A0A1B0ANU1 A0A1L8F0A6 A0A1V4KZW4 T1E1S1 E1C5K5 A0A226N981 A0A1I8MCK0 F6QN31 A0A1A9X053 A0A2K5MNC2 A0A1A9Y9D1 A0A093GBR7 A0A1I8P510 A0A3P8YXR4 A0A250XYX8 W8BLI5 A0A1L8F9V6 A0A226NZN2 A0A1I8P562 A0A0L0BX11 Q0IFK7 A0A084WE48 A0A250XZA3 A0A182J1V0 A0A2K5IJJ4 A0A2J8WKH2 A0A2K6D4B2 A0A2K5WC89 A0A2K5YIX4 F7H9C7 A0A2J8WKG5 A0A2R9C6X5 H2Q899 A0A182NFT3 G3RYF2 Q00532-3
A0A212FPW4 D6WSF5 A0A158NAV6 A0A232EVU3 A0A0K8TJ02 A0A067R7U0 A0A2P8XRW4 A0A026WUU7 A0A2H8TLW9 E2AT42 A0A2S2NVL7 A0A1S4F8D7 A0A1B6CAC5 J9JXR5 E0VSK6 X2JD86 A0A0J9QWB6 A0A0P5K232 A0A0P5A5F0 A0A0P5CLC3 A0A182QK93 A0A0R3NZS1 A0A0P5JNY3 A0A1W4VBS3 A0A0P6AH56 A0A0P6GUD6 A0A2A3EJD4 A0A0N8EN14 A0A3L8D8P0 A0A182R375 A0A182XE57 U4U3Z9 A0A1Y1LJJ0 A0A1D2NE10 K7J404 N6TBM8 A0A2R7WIC0 A0A0K8TIH2 A0A310SXL0 A0A154P6C2 V9LIN5 F4X245 A0A195E4V6 A0A151I5W7 A0A0L7QK80 A0A195EVD1 A0A195CK25 E9G338 A0A1A9USN7 E2BPF6 A0A088ABR9 A0A0M9A4V5 A0A034V2J7 A0A2P6L9K2 A0A087UTB1 A0A226DFG1 A0A182H5B4 A0A1B0G098 A0A1B0ANU1 A0A1L8F0A6 A0A1V4KZW4 T1E1S1 E1C5K5 A0A226N981 A0A1I8MCK0 F6QN31 A0A1A9X053 A0A2K5MNC2 A0A1A9Y9D1 A0A093GBR7 A0A1I8P510 A0A3P8YXR4 A0A250XYX8 W8BLI5 A0A1L8F9V6 A0A226NZN2 A0A1I8P562 A0A0L0BX11 Q0IFK7 A0A084WE48 A0A250XZA3 A0A182J1V0 A0A2K5IJJ4 A0A2J8WKH2 A0A2K6D4B2 A0A2K5WC89 A0A2K5YIX4 F7H9C7 A0A2J8WKG5 A0A2R9C6X5 H2Q899 A0A182NFT3 G3RYF2 Q00532-3
EC Number
2.7.11.22
Pubmed
26354079
26227816
19121390
22118469
18362917
19820115
+ More
21347285 28648823 24845553 29403074 24508170 20798317 20566863 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 22936249 15632085 17994087 30249741 23537049 28004739 27289101 20075255 24402279 21719571 21292972 25348373 26483478 27762356 15592404 25315136 20431018 25069045 28087693 24495485 24621616 26108605 17510324 24438588 17431167 22722832 16136131 22398555 1639063 12508121 14702039 15489334 9000130 29420175 17344846
21347285 28648823 24845553 29403074 24508170 20798317 20566863 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 22936249 15632085 17994087 30249741 23537049 28004739 27289101 20075255 24402279 21719571 21292972 25348373 26483478 27762356 15592404 25315136 20431018 25069045 28087693 24495485 24621616 26108605 17510324 24438588 17431167 22722832 16136131 22398555 1639063 12508121 14702039 15489334 9000130 29420175 17344846
EMBL
ODYU01011565
SOQ57382.1
NWSH01000577
PCG75539.1
RSAL01000009
RVE53868.1
+ More
KQ459604 KPI92064.1 JTDY01001012 KOB75129.1 BABH01025204 BABH01025205 BABH01025206 AGBW02000220 OWR55802.1 KQ971352 EFA06376.2 ADTU01010316 ADTU01010317 NNAY01001957 OXU22456.1 GBRD01000415 JAG65406.1 KK852683 KDR18537.1 PYGN01001453 PSN34748.1 KK107105 EZA59436.1 GFXV01003214 MBW15019.1 GL442489 EFN63396.1 GGMR01008620 MBY21239.1 GEDC01027098 JAS10200.1 ABLF02020383 ABLF02020384 ABLF02038505 ABLF02038519 ABLF02057788 DS235751 EEB16362.1 AE014134 AHN54150.1 CM002910 KMY88350.1 KMY88351.1 GDIQ01195791 JAK55934.1 GDIP01208232 JAJ15170.1 GDIP01168895 JAJ54507.1 AXCN02000244 CH379061 KRT04765.1 GDIQ01197341 JAK54384.1 GDIP01030816 JAM72899.1 GDIQ01029523 LRGB01000512 JAN65214.1 KZS18439.1 KZ288239 PBC31309.1 GDIQ01010756 JAN83981.1 QOIP01000011 RLU16887.1 KB632083 ERL88619.1 GEZM01055704 JAV72978.1 LJIJ01000071 ODN03489.1 AAZX01001010 APGK01036456 KB740940 ENN77679.1 KK854877 PTY19394.1 GBRD01000414 JAG65407.1 KQ759796 OAD62826.1 KQ434809 KZC06690.1 JW880723 AFP13240.1 GL888565 EGI59461.1 KQ979609 KYN20220.1 KQ976413 KYM90415.1 KQ414983 KOC59014.1 KQ981954 KYN32220.1 KQ977642 KYN01085.1 GL732530 EFX86396.1 GL449633 EFN82360.1 KQ435733 KOX77277.1 GAKP01023193 JAC35763.1 MWRG01000796 PRD35248.1 KK121490 KFM80600.1 LNIX01000022 OXA43447.1 JXUM01111200 JXUM01111201 KQ565480 KXJ70927.1 CCAG010018487 JXJN01001010 JXJN01001011 CM004481 OCT65003.1 LSYS01000634 OPJ90029.1 GAKT01000040 JAA93022.1 AADN05000303 MCFN01000133 OXB64058.1 AAMC01077504 AAMC01077505 KL215798 KFV66633.1 GFFV01003381 JAV36564.1 GAMC01016056 JAB90499.1 CM004480 OCT68318.1 AWGT02000251 OXB72810.1 JRES01001211 KNC24550.1 CH477312 EAT43896.1 ATLV01023139 KE525341 KFB48492.1 GFFV01003320 JAV36625.1 NDHI03003386 PNJ70268.1 AQIA01064846 AQIA01064847 JSUE03039704 PNJ70265.1 AJFE02112170 AJFE02112171 AJFE02112172 AJFE02112173 AJFE02112174 AACZ04008568 NBAG03000214 PNI83323.1 CABD030092265 CABD030092266 CABD030092267 X66358 X66359 AF390028 AY525548 AL118556 AL359397 AK308732 BC104977
KQ459604 KPI92064.1 JTDY01001012 KOB75129.1 BABH01025204 BABH01025205 BABH01025206 AGBW02000220 OWR55802.1 KQ971352 EFA06376.2 ADTU01010316 ADTU01010317 NNAY01001957 OXU22456.1 GBRD01000415 JAG65406.1 KK852683 KDR18537.1 PYGN01001453 PSN34748.1 KK107105 EZA59436.1 GFXV01003214 MBW15019.1 GL442489 EFN63396.1 GGMR01008620 MBY21239.1 GEDC01027098 JAS10200.1 ABLF02020383 ABLF02020384 ABLF02038505 ABLF02038519 ABLF02057788 DS235751 EEB16362.1 AE014134 AHN54150.1 CM002910 KMY88350.1 KMY88351.1 GDIQ01195791 JAK55934.1 GDIP01208232 JAJ15170.1 GDIP01168895 JAJ54507.1 AXCN02000244 CH379061 KRT04765.1 GDIQ01197341 JAK54384.1 GDIP01030816 JAM72899.1 GDIQ01029523 LRGB01000512 JAN65214.1 KZS18439.1 KZ288239 PBC31309.1 GDIQ01010756 JAN83981.1 QOIP01000011 RLU16887.1 KB632083 ERL88619.1 GEZM01055704 JAV72978.1 LJIJ01000071 ODN03489.1 AAZX01001010 APGK01036456 KB740940 ENN77679.1 KK854877 PTY19394.1 GBRD01000414 JAG65407.1 KQ759796 OAD62826.1 KQ434809 KZC06690.1 JW880723 AFP13240.1 GL888565 EGI59461.1 KQ979609 KYN20220.1 KQ976413 KYM90415.1 KQ414983 KOC59014.1 KQ981954 KYN32220.1 KQ977642 KYN01085.1 GL732530 EFX86396.1 GL449633 EFN82360.1 KQ435733 KOX77277.1 GAKP01023193 JAC35763.1 MWRG01000796 PRD35248.1 KK121490 KFM80600.1 LNIX01000022 OXA43447.1 JXUM01111200 JXUM01111201 KQ565480 KXJ70927.1 CCAG010018487 JXJN01001010 JXJN01001011 CM004481 OCT65003.1 LSYS01000634 OPJ90029.1 GAKT01000040 JAA93022.1 AADN05000303 MCFN01000133 OXB64058.1 AAMC01077504 AAMC01077505 KL215798 KFV66633.1 GFFV01003381 JAV36564.1 GAMC01016056 JAB90499.1 CM004480 OCT68318.1 AWGT02000251 OXB72810.1 JRES01001211 KNC24550.1 CH477312 EAT43896.1 ATLV01023139 KE525341 KFB48492.1 GFFV01003320 JAV36625.1 NDHI03003386 PNJ70268.1 AQIA01064846 AQIA01064847 JSUE03039704 PNJ70265.1 AJFE02112170 AJFE02112171 AJFE02112172 AJFE02112173 AJFE02112174 AACZ04008568 NBAG03000214 PNI83323.1 CABD030092265 CABD030092266 CABD030092267 X66358 X66359 AF390028 AY525548 AL118556 AL359397 AK308732 BC104977
Proteomes
UP000218220
UP000283053
UP000053268
UP000037510
UP000005204
UP000007151
+ More
UP000007266 UP000005205 UP000215335 UP000027135 UP000245037 UP000053097 UP000000311 UP000007819 UP000009046 UP000000803 UP000075886 UP000001819 UP000192221 UP000076858 UP000242457 UP000279307 UP000075900 UP000076407 UP000030742 UP000094527 UP000002358 UP000019118 UP000076502 UP000007755 UP000078492 UP000078540 UP000053825 UP000078541 UP000078542 UP000000305 UP000078200 UP000008237 UP000005203 UP000053105 UP000054359 UP000198287 UP000069940 UP000249989 UP000092444 UP000092460 UP000186698 UP000190648 UP000000539 UP000198323 UP000095301 UP000008143 UP000091820 UP000233060 UP000092443 UP000053875 UP000095300 UP000265140 UP000198419 UP000037069 UP000008820 UP000030765 UP000075880 UP000233080 UP000233120 UP000233100 UP000233140 UP000006718 UP000240080 UP000002277 UP000075884 UP000001519 UP000005640
UP000007266 UP000005205 UP000215335 UP000027135 UP000245037 UP000053097 UP000000311 UP000007819 UP000009046 UP000000803 UP000075886 UP000001819 UP000192221 UP000076858 UP000242457 UP000279307 UP000075900 UP000076407 UP000030742 UP000094527 UP000002358 UP000019118 UP000076502 UP000007755 UP000078492 UP000078540 UP000053825 UP000078541 UP000078542 UP000000305 UP000078200 UP000008237 UP000005203 UP000053105 UP000054359 UP000198287 UP000069940 UP000249989 UP000092444 UP000092460 UP000186698 UP000190648 UP000000539 UP000198323 UP000095301 UP000008143 UP000091820 UP000233060 UP000092443 UP000053875 UP000095300 UP000265140 UP000198419 UP000037069 UP000008820 UP000030765 UP000075880 UP000233080 UP000233120 UP000233100 UP000233140 UP000006718 UP000240080 UP000002277 UP000075884 UP000001519 UP000005640
Interpro
Gene 3D
ProteinModelPortal
A0A2H1WWL6
A0A2A4JUQ8
A0A3S2LHD6
A0A194PH83
A0A0L7LID4
H9J5K0
+ More
A0A212FPW4 D6WSF5 A0A158NAV6 A0A232EVU3 A0A0K8TJ02 A0A067R7U0 A0A2P8XRW4 A0A026WUU7 A0A2H8TLW9 E2AT42 A0A2S2NVL7 A0A1S4F8D7 A0A1B6CAC5 J9JXR5 E0VSK6 X2JD86 A0A0J9QWB6 A0A0P5K232 A0A0P5A5F0 A0A0P5CLC3 A0A182QK93 A0A0R3NZS1 A0A0P5JNY3 A0A1W4VBS3 A0A0P6AH56 A0A0P6GUD6 A0A2A3EJD4 A0A0N8EN14 A0A3L8D8P0 A0A182R375 A0A182XE57 U4U3Z9 A0A1Y1LJJ0 A0A1D2NE10 K7J404 N6TBM8 A0A2R7WIC0 A0A0K8TIH2 A0A310SXL0 A0A154P6C2 V9LIN5 F4X245 A0A195E4V6 A0A151I5W7 A0A0L7QK80 A0A195EVD1 A0A195CK25 E9G338 A0A1A9USN7 E2BPF6 A0A088ABR9 A0A0M9A4V5 A0A034V2J7 A0A2P6L9K2 A0A087UTB1 A0A226DFG1 A0A182H5B4 A0A1B0G098 A0A1B0ANU1 A0A1L8F0A6 A0A1V4KZW4 T1E1S1 E1C5K5 A0A226N981 A0A1I8MCK0 F6QN31 A0A1A9X053 A0A2K5MNC2 A0A1A9Y9D1 A0A093GBR7 A0A1I8P510 A0A3P8YXR4 A0A250XYX8 W8BLI5 A0A1L8F9V6 A0A226NZN2 A0A1I8P562 A0A0L0BX11 Q0IFK7 A0A084WE48 A0A250XZA3 A0A182J1V0 A0A2K5IJJ4 A0A2J8WKH2 A0A2K6D4B2 A0A2K5WC89 A0A2K5YIX4 F7H9C7 A0A2J8WKG5 A0A2R9C6X5 H2Q899 A0A182NFT3 G3RYF2 Q00532-3
A0A212FPW4 D6WSF5 A0A158NAV6 A0A232EVU3 A0A0K8TJ02 A0A067R7U0 A0A2P8XRW4 A0A026WUU7 A0A2H8TLW9 E2AT42 A0A2S2NVL7 A0A1S4F8D7 A0A1B6CAC5 J9JXR5 E0VSK6 X2JD86 A0A0J9QWB6 A0A0P5K232 A0A0P5A5F0 A0A0P5CLC3 A0A182QK93 A0A0R3NZS1 A0A0P5JNY3 A0A1W4VBS3 A0A0P6AH56 A0A0P6GUD6 A0A2A3EJD4 A0A0N8EN14 A0A3L8D8P0 A0A182R375 A0A182XE57 U4U3Z9 A0A1Y1LJJ0 A0A1D2NE10 K7J404 N6TBM8 A0A2R7WIC0 A0A0K8TIH2 A0A310SXL0 A0A154P6C2 V9LIN5 F4X245 A0A195E4V6 A0A151I5W7 A0A0L7QK80 A0A195EVD1 A0A195CK25 E9G338 A0A1A9USN7 E2BPF6 A0A088ABR9 A0A0M9A4V5 A0A034V2J7 A0A2P6L9K2 A0A087UTB1 A0A226DFG1 A0A182H5B4 A0A1B0G098 A0A1B0ANU1 A0A1L8F0A6 A0A1V4KZW4 T1E1S1 E1C5K5 A0A226N981 A0A1I8MCK0 F6QN31 A0A1A9X053 A0A2K5MNC2 A0A1A9Y9D1 A0A093GBR7 A0A1I8P510 A0A3P8YXR4 A0A250XYX8 W8BLI5 A0A1L8F9V6 A0A226NZN2 A0A1I8P562 A0A0L0BX11 Q0IFK7 A0A084WE48 A0A250XZA3 A0A182J1V0 A0A2K5IJJ4 A0A2J8WKH2 A0A2K6D4B2 A0A2K5WC89 A0A2K5YIX4 F7H9C7 A0A2J8WKG5 A0A2R9C6X5 H2Q899 A0A182NFT3 G3RYF2 Q00532-3
PDB
4AGU
E-value=9.64519e-71,
Score=675
Ontologies
GO
PANTHER
Topology
Subcellular location
Cytoplasm
Nucleus
Nucleus
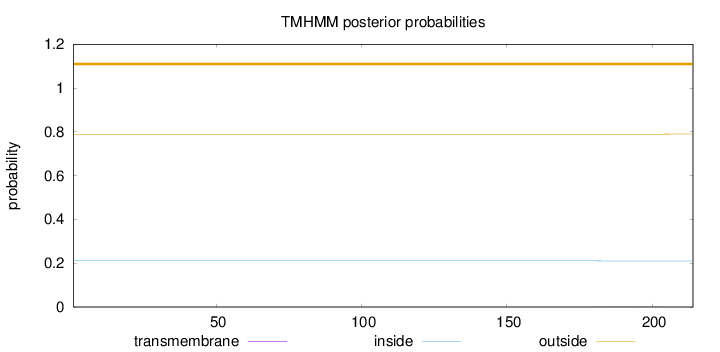
Length:
214
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.02927
Exp number, first 60 AAs:
0.00116
Total prob of N-in:
0.21118
outside
1 - 214
Population Genetic Test Statistics
Pi
185.037024
Theta
173.597671
Tajima's D
0.233605
CLR
1.164633
CSRT
0.43832808359582
Interpretation
Uncertain
