Gene
KWMTBOMO14990
Pre Gene Modal
BGIBMGA004784
Annotation
PREDICTED:_protein_CLP1_homolog_[Amyelois_transitella]
Full name
Protein CLP1 homolog
+ More
Polyribonucleotide 5'-hydroxyl-kinase Clp1
Polyribonucleotide 5'-hydroxyl-kinase Clp1
Alternative Name
Polyadenylation factor Clp1
Polynucleotide kinase Clp1
Pre-mRNA cleavage complex II protein Clp1
Polynucleotide kinase Clp1
Pre-mRNA cleavage complex II protein Clp1
Location in the cell
Cytoplasmic Reliability : 2.955
Sequence
CDS
ATGACTGAAGTGCAACTACAAGAATTTAAGCTTGACCCCGATTCGGAATTACGTTTTGAAGTAGAATCGAAGAACGAAAAAGTCGTCTTAGAGGTAAAAAGTGGATATGCTGAATTATTCGGTACAGAACTGGTGAAAGGAAAGCCATACGAATTTCACACAGGAGCTAAAGTTGCTGTGTTTACTTGGCACGGATGTACTATAGAACTCAGAGGAAAGACTGAAGTCAGCTATGTAGCCAAAGAAACACCGATGATAGTGTACTTGAACGTACATGCGGCATTAGAACAGCAAAGAGTTGCAGCAGAACAAGAGAACACAAGAGGTCCGGTAACAATGATAGTTGGGCCTGCAGATGTTGGCAAGTCCACACTCTCAAGGATCCTTTTGAACTATGCTGTCCGCATGGGCAGGAGGCCTATTTATGTTGATTTGGATGTGGGACAGGGCCAGATTAGCGTTCCTGGTACAATCGGTGCATTGTTAGTAGAGCGCCCTGCGTCTATAGAAGAAGGCTTCAGTCAGCAGGCTCCGTTGGTTTATCATTTTGGTCACACTTCGCCGAGCGATAATATTGAGTTATACAACACCATTGTGTCGAGGCTAGCCGAAGTGATCACAGAGAAATGCGAAACGAATAAAAAAGCGGCGTCTTCGGGCGTCATAATAAACACGTGTGGATGGATTAAGAATGCTGGTTACAAGGTTTTAACTCATGCCGCTCAAGCATTTGAGGTTGACGTCATATTAGTTTTAGACAATGAGAGATTGTACAACGAGCTGAAGCGTGACATGCCTAAATTTGTGAAAGTAGTCTATCTACCTAAAAGTGGTGGGGTGGTGGAGCGCTCCGGCACGCAGAGGGCGGAGGCCCGGGACGCGCGCATCCGGGAGTACTTCTACGGGAAGAGAACACCATACTACCCGCATTCCTTTGATGTTAAATTCTCAGATCTCAAGATATACAAGGTGGGCGCGCCGTCGCTGCCGGACTCGTGCATGCCGCTGGGGATGCGCGCGGAGGACGCGCTGACGCGGCTGGTGGCGGTGTGGGGGGGCCGCGCGCTGCAGCACCGCCTGCTCGCCGTGTCCTTCGCGGCGGAGCCCGACGACCACGTGCTGCACTCCAACCTCGCGGGGTTCGTCTGTGTAACGGCAGTCGACATGGAGCGACAGACTTTGACGGTCCTGTCCCCGCAGCCGCGCCCTCTGCCCAACACGATCCTGTTGTTATCAGAACTACAATACATGGACAATCATTAG
Protein
MTEVQLQEFKLDPDSELRFEVESKNEKVVLEVKSGYAELFGTELVKGKPYEFHTGAKVAVFTWHGCTIELRGKTEVSYVAKETPMIVYLNVHAALEQQRVAAEQENTRGPVTMIVGPADVGKSTLSRILLNYAVRMGRRPIYVDLDVGQGQISVPGTIGALLVERPASIEEGFSQQAPLVYHFGHTSPSDNIELYNTIVSRLAEVITEKCETNKKAASSGVIINTCGWIKNAGYKVLTHAAQAFEVDVILVLDNERLYNELKRDMPKFVKVVYLPKSGGVVERSGTQRAEARDARIREYFYGKRTPYYPHSFDVKFSDLKIYKVGAPSLPDSCMPLGMRAEDALTRLVAVWGGRALQHRLLAVSFAAEPDDHVLHSNLAGFVCVTAVDMERQTLTVLSPQPRPLPNTILLLSELQYMDNH
Summary
Description
Required for endonucleolytic cleavage during polyadenylation-dependent pre-mRNA 3'-end formation.
Polynucleotide kinase that can phosphorylate the 5'-hydroxyl groups of double-stranded RNA (dsRNA), single-stranded RNA (ssRNA), double stranded DNA (dsDNA) and double-stranded DNA:RNA hybrids. dsRNA is phosphorylated more efficiently than dsDNA, and the RNA component of a DNA:RNA hybrid is phosphorylated more efficiently than the DNA component. Appears to have roles in both tRNA splicing and mRNA 3'-end formation. Component of the tRNA splicing endonuclease complex. Phosphorylates the 5'-terminus of the tRNA 3'-exon during tRNA splicing; this phosphorylation event is a prerequisite for the subsequent ligation of the two exon halves and the production of a mature tRNA. Component of the pre-mRNA cleavage complex II (CF-II), which seems to be required for mRNA 3'-end formation. Also phosphorylates the 5'-terminus of exogenously introduced short interfering RNAs (siRNAs), which is a necessary prerequisite for their incorporation into the RNA-induced silencing complex (RISC). However, endogenous siRNAs and microRNAs (miRNAs) that are produced by the cleavage of dsRNA precursors by DICER1 already contain a 5'-phosphate group, so this protein may be dispensible for normal RNA-mediated gene silencing.
Polynucleotide kinase that can phosphorylate the 5'-hydroxyl groups of double-stranded RNA (dsRNA), single-stranded RNA (ssRNA), double stranded DNA (dsDNA) and double-stranded DNA:RNA hybrids. dsRNA is phosphorylated more efficiently than dsDNA, and the RNA component of a DNA:RNA hybrid is phosphorylated more efficiently than the DNA component. Appears to have roles in both tRNA splicing and mRNA 3'-end formation. Component of the tRNA splicing endonuclease complex. Phosphorylates the 5'-terminus of the tRNA 3'-exon during tRNA splicing; this phosphorylation event is a prerequisite for the subsequent ligation of the two exon halves and the production of a mature tRNA. Component of the pre-mRNA cleavage complex II (CF-II), which seems to be required for mRNA 3'-end formation. Also phosphorylates the 5'-terminus of exogenously introduced short interfering RNAs (siRNAs), which is a necessary prerequisite for their incorporation into the RNA-induced silencing complex (RISC). However, endogenous siRNAs and microRNAs (miRNAs) that are produced by the cleavage of dsRNA precursors by DICER1 already contain a 5'-phosphate group, so this protein may be dispensible for normal RNA-mediated gene silencing.
Catalytic Activity
5'-dephospho-(DNA) + ATP = ADP + H(+) + phospho-5'-(DNA)
5'-dephospho-(RNA) + ATP = 5'-end-(RNA) + ADP + H(+)
5'-dephospho-(RNA) + ATP = 5'-end-(RNA) + ADP + H(+)
Subunit
Component of the tRNA splicing endonuclease complex. Component of pre-mRNA cleavage complex II (CF-II).
Similarity
Belongs to the Clp1 family. Clp1 subfamily.
Keywords
ATP-binding
Complete proteome
mRNA processing
Nucleotide-binding
Nucleus
Reference proteome
Feature
chain Protein CLP1 homolog
Uniprot
S4PM57
A0A2A4JSV0
A0A2H1V530
A0A194RI29
A0A194PH92
H9J5J4
+ More
A0A212EUZ5 A0A067QNZ6 A0A2J7Q9P9 A0A3L8DBP0 A0A195FIC7 F4W693 A0A026WFV2 A0A151WHS8 A0A195AVL0 A0A158P214 A0A2A3E1S2 A0A088AW37 E9ISC1 A0A1B6EEL2 A0A0J7L6Z8 E2BIM6 A0A1B6BXL4 A0A1B6GLU8 A0A0C9R7A9 K7J1J2 A0A232FIT2 V5GVE7 A0A1E1X2W4 G3MND4 A0A023GK14 A0A131XH63 L7M8T0 A0A224YLY6 A0A131YSF0 A0A1Z5L6P2 A0A293MCY9 A0A1B0CM02 A0A1B0D1L0 A0A2R5LJW0 A0A0A9W5J4 A0A139WEM6 A0A1W4WBN3 A0A1Y1L955 E0VBS2 A0A0P4W2R7 A0A023F4J3 A0A224XPF8 A0A2R7VRS5 A0A069DU68 A0A182PA03 A0A182M913 B0VZR4 A0A0V0G7N5 A0A182WIY4 A0A182QEX3 A0A1Q3FCE4 A0A182RB20 A0A336M572 A0A182XZC6 A0A182UTN6 A0A336MQF3 A0A1C7CZJ7 A0A182UCF7 A0A182X6K8 A0A182I3V7 Q7QJW7 A0A182NHC8 U5EVR2 A0A182JCE3 A0A084VF65 A0A182JTV8 A0A2M4CTH4 A0A336MN00 A0A0K8WG81 A0A182FG29 A0A1B6EA97 A0A1A9W0M8 A0A0P5ZAU3 A0A034W9A0 A0A0A1WL03 A0A0N0BBL1 W8C415 D3TPC4 A0A1I8NFB6 A0A0L0BXL2 T1J8D4 E9FU50 Q16WA6 A0A2K7P6C0 A0A1I8QAR4 A0A1B0G3E5 A0A1D2NCD2 A0A1L8DGL6 A0A0P6IHX3 A0A087TNZ7 A0A182H1F3 A0A2I0TWQ1 G1NBF2 A0A226MWM2
A0A212EUZ5 A0A067QNZ6 A0A2J7Q9P9 A0A3L8DBP0 A0A195FIC7 F4W693 A0A026WFV2 A0A151WHS8 A0A195AVL0 A0A158P214 A0A2A3E1S2 A0A088AW37 E9ISC1 A0A1B6EEL2 A0A0J7L6Z8 E2BIM6 A0A1B6BXL4 A0A1B6GLU8 A0A0C9R7A9 K7J1J2 A0A232FIT2 V5GVE7 A0A1E1X2W4 G3MND4 A0A023GK14 A0A131XH63 L7M8T0 A0A224YLY6 A0A131YSF0 A0A1Z5L6P2 A0A293MCY9 A0A1B0CM02 A0A1B0D1L0 A0A2R5LJW0 A0A0A9W5J4 A0A139WEM6 A0A1W4WBN3 A0A1Y1L955 E0VBS2 A0A0P4W2R7 A0A023F4J3 A0A224XPF8 A0A2R7VRS5 A0A069DU68 A0A182PA03 A0A182M913 B0VZR4 A0A0V0G7N5 A0A182WIY4 A0A182QEX3 A0A1Q3FCE4 A0A182RB20 A0A336M572 A0A182XZC6 A0A182UTN6 A0A336MQF3 A0A1C7CZJ7 A0A182UCF7 A0A182X6K8 A0A182I3V7 Q7QJW7 A0A182NHC8 U5EVR2 A0A182JCE3 A0A084VF65 A0A182JTV8 A0A2M4CTH4 A0A336MN00 A0A0K8WG81 A0A182FG29 A0A1B6EA97 A0A1A9W0M8 A0A0P5ZAU3 A0A034W9A0 A0A0A1WL03 A0A0N0BBL1 W8C415 D3TPC4 A0A1I8NFB6 A0A0L0BXL2 T1J8D4 E9FU50 Q16WA6 A0A2K7P6C0 A0A1I8QAR4 A0A1B0G3E5 A0A1D2NCD2 A0A1L8DGL6 A0A0P6IHX3 A0A087TNZ7 A0A182H1F3 A0A2I0TWQ1 G1NBF2 A0A226MWM2
EC Number
2.7.1.78
Pubmed
23622113
26354079
19121390
22118469
24845553
30249741
+ More
21719571 24508170 21347285 21282665 20798317 20075255 28648823 25765539 28503490 22216098 28049606 25576852 28797301 26830274 28528879 25401762 26823975 18362917 19820115 28004739 20566863 27129103 25474469 26334808 25244985 20966253 12364791 24438588 25348373 25830018 24495485 20353571 25315136 26108605 21292972 17510324 27289101 26483478 20838655
21719571 24508170 21347285 21282665 20798317 20075255 28648823 25765539 28503490 22216098 28049606 25576852 28797301 26830274 28528879 25401762 26823975 18362917 19820115 28004739 20566863 27129103 25474469 26334808 25244985 20966253 12364791 24438588 25348373 25830018 24495485 20353571 25315136 26108605 21292972 17510324 27289101 26483478 20838655
EMBL
GAIX01000291
JAA92269.1
NWSH01000636
PCG75117.1
ODYU01000712
SOQ35941.1
+ More
KQ460205 KPJ16985.1 KQ459604 KPI92074.1 BABH01025225 AGBW02012265 OWR45315.1 KK853123 KDR11058.1 NEVH01016344 PNF25313.1 QOIP01000010 RLU17299.1 KQ981523 KYN40425.1 GL887707 EGI70228.1 KK107262 EZA53904.1 KQ983112 KYQ47368.1 KQ976731 KYM76273.1 ADTU01006959 KZ288490 PBC25206.1 GL765324 EFZ16539.1 GEDC01000957 JAS36341.1 LBMM01000338 KMR00323.1 GL448516 EFN84473.1 GEDC01031548 JAS05750.1 GECZ01006371 JAS63398.1 GBYB01002641 JAG72408.1 NNAY01000155 OXU30493.1 GANP01010103 JAB74365.1 GFAC01005568 JAT93620.1 JO843385 AEO35002.1 GBBM01001187 JAC34231.1 GEFH01003565 JAP65016.1 GACK01004762 JAA60272.1 GFPF01006819 MAA17965.1 GEDV01007039 JAP81518.1 GFJQ02004246 JAW02724.1 GFWV01012281 MAA37010.1 AJWK01017982 AJVK01022110 GGLE01005706 MBY09832.1 GBHO01039892 GBRD01012824 GDHC01004401 JAG03712.1 JAG53002.1 JAQ14228.1 KQ971354 KYB26428.1 GEZM01066959 GEZM01066958 GEZM01066957 GEZM01066956 JAV67617.1 DS235035 EEB10828.1 GDKW01000792 JAI55803.1 GBBI01002565 JAC16147.1 GFTR01006106 JAW10320.1 KK854031 PTY09848.1 GBGD01001578 JAC87311.1 AXCM01001459 DS231815 GECL01002197 JAP03927.1 AXCN02000896 GFDL01009799 JAV25246.1 UFQS01000450 UFQT01000450 SSX04136.1 SSX24501.1 UFQT01001558 SSX31013.1 APCN01000225 AAAB01008807 GANO01000905 JAB58966.1 ATLV01012362 KE524785 KFB36609.1 GGFL01004401 MBW68579.1 SSX31011.1 GDHF01002217 JAI50097.1 GEDC01025302 GEDC01002461 JAS11996.1 JAS34837.1 GDIP01046258 LRGB01001937 JAM57457.1 KZS09962.1 GAKP01008050 JAC50902.1 GBXI01015092 JAC99199.1 KQ436240 KOX67341.1 GAMC01002537 JAC04019.1 EZ423276 ADD19552.1 JRES01001180 KNC24736.1 JH431954 GL732524 EFX89485.1 CH477572 CCAG010009527 LJIJ01000093 ODN02899.1 GFDF01008600 JAV05484.1 GDIQ01021713 JAN73024.1 KK116106 KFM66836.1 JXUM01023000 KQ560647 KXJ81453.1 KZ506809 PKU38237.1 MCFN01000375 OXB59727.1
KQ460205 KPJ16985.1 KQ459604 KPI92074.1 BABH01025225 AGBW02012265 OWR45315.1 KK853123 KDR11058.1 NEVH01016344 PNF25313.1 QOIP01000010 RLU17299.1 KQ981523 KYN40425.1 GL887707 EGI70228.1 KK107262 EZA53904.1 KQ983112 KYQ47368.1 KQ976731 KYM76273.1 ADTU01006959 KZ288490 PBC25206.1 GL765324 EFZ16539.1 GEDC01000957 JAS36341.1 LBMM01000338 KMR00323.1 GL448516 EFN84473.1 GEDC01031548 JAS05750.1 GECZ01006371 JAS63398.1 GBYB01002641 JAG72408.1 NNAY01000155 OXU30493.1 GANP01010103 JAB74365.1 GFAC01005568 JAT93620.1 JO843385 AEO35002.1 GBBM01001187 JAC34231.1 GEFH01003565 JAP65016.1 GACK01004762 JAA60272.1 GFPF01006819 MAA17965.1 GEDV01007039 JAP81518.1 GFJQ02004246 JAW02724.1 GFWV01012281 MAA37010.1 AJWK01017982 AJVK01022110 GGLE01005706 MBY09832.1 GBHO01039892 GBRD01012824 GDHC01004401 JAG03712.1 JAG53002.1 JAQ14228.1 KQ971354 KYB26428.1 GEZM01066959 GEZM01066958 GEZM01066957 GEZM01066956 JAV67617.1 DS235035 EEB10828.1 GDKW01000792 JAI55803.1 GBBI01002565 JAC16147.1 GFTR01006106 JAW10320.1 KK854031 PTY09848.1 GBGD01001578 JAC87311.1 AXCM01001459 DS231815 GECL01002197 JAP03927.1 AXCN02000896 GFDL01009799 JAV25246.1 UFQS01000450 UFQT01000450 SSX04136.1 SSX24501.1 UFQT01001558 SSX31013.1 APCN01000225 AAAB01008807 GANO01000905 JAB58966.1 ATLV01012362 KE524785 KFB36609.1 GGFL01004401 MBW68579.1 SSX31011.1 GDHF01002217 JAI50097.1 GEDC01025302 GEDC01002461 JAS11996.1 JAS34837.1 GDIP01046258 LRGB01001937 JAM57457.1 KZS09962.1 GAKP01008050 JAC50902.1 GBXI01015092 JAC99199.1 KQ436240 KOX67341.1 GAMC01002537 JAC04019.1 EZ423276 ADD19552.1 JRES01001180 KNC24736.1 JH431954 GL732524 EFX89485.1 CH477572 CCAG010009527 LJIJ01000093 ODN02899.1 GFDF01008600 JAV05484.1 GDIQ01021713 JAN73024.1 KK116106 KFM66836.1 JXUM01023000 KQ560647 KXJ81453.1 KZ506809 PKU38237.1 MCFN01000375 OXB59727.1
Proteomes
UP000218220
UP000053240
UP000053268
UP000005204
UP000007151
UP000027135
+ More
UP000235965 UP000279307 UP000078541 UP000007755 UP000053097 UP000075809 UP000078540 UP000005205 UP000242457 UP000005203 UP000036403 UP000008237 UP000002358 UP000215335 UP000092461 UP000092462 UP000007266 UP000192223 UP000009046 UP000075885 UP000075883 UP000002320 UP000075920 UP000075886 UP000075900 UP000076408 UP000075903 UP000075882 UP000075902 UP000076407 UP000075840 UP000007062 UP000075884 UP000075880 UP000030765 UP000075881 UP000069272 UP000091820 UP000076858 UP000053105 UP000095301 UP000037069 UP000000305 UP000008820 UP000095300 UP000092444 UP000094527 UP000054359 UP000069940 UP000249989 UP000001645 UP000198323
UP000235965 UP000279307 UP000078541 UP000007755 UP000053097 UP000075809 UP000078540 UP000005205 UP000242457 UP000005203 UP000036403 UP000008237 UP000002358 UP000215335 UP000092461 UP000092462 UP000007266 UP000192223 UP000009046 UP000075885 UP000075883 UP000002320 UP000075920 UP000075886 UP000075900 UP000076408 UP000075903 UP000075882 UP000075902 UP000076407 UP000075840 UP000007062 UP000075884 UP000075880 UP000030765 UP000075881 UP000069272 UP000091820 UP000076858 UP000053105 UP000095301 UP000037069 UP000000305 UP000008820 UP000095300 UP000092444 UP000094527 UP000054359 UP000069940 UP000249989 UP000001645 UP000198323
PRIDE
Interpro
ProteinModelPortal
S4PM57
A0A2A4JSV0
A0A2H1V530
A0A194RI29
A0A194PH92
H9J5J4
+ More
A0A212EUZ5 A0A067QNZ6 A0A2J7Q9P9 A0A3L8DBP0 A0A195FIC7 F4W693 A0A026WFV2 A0A151WHS8 A0A195AVL0 A0A158P214 A0A2A3E1S2 A0A088AW37 E9ISC1 A0A1B6EEL2 A0A0J7L6Z8 E2BIM6 A0A1B6BXL4 A0A1B6GLU8 A0A0C9R7A9 K7J1J2 A0A232FIT2 V5GVE7 A0A1E1X2W4 G3MND4 A0A023GK14 A0A131XH63 L7M8T0 A0A224YLY6 A0A131YSF0 A0A1Z5L6P2 A0A293MCY9 A0A1B0CM02 A0A1B0D1L0 A0A2R5LJW0 A0A0A9W5J4 A0A139WEM6 A0A1W4WBN3 A0A1Y1L955 E0VBS2 A0A0P4W2R7 A0A023F4J3 A0A224XPF8 A0A2R7VRS5 A0A069DU68 A0A182PA03 A0A182M913 B0VZR4 A0A0V0G7N5 A0A182WIY4 A0A182QEX3 A0A1Q3FCE4 A0A182RB20 A0A336M572 A0A182XZC6 A0A182UTN6 A0A336MQF3 A0A1C7CZJ7 A0A182UCF7 A0A182X6K8 A0A182I3V7 Q7QJW7 A0A182NHC8 U5EVR2 A0A182JCE3 A0A084VF65 A0A182JTV8 A0A2M4CTH4 A0A336MN00 A0A0K8WG81 A0A182FG29 A0A1B6EA97 A0A1A9W0M8 A0A0P5ZAU3 A0A034W9A0 A0A0A1WL03 A0A0N0BBL1 W8C415 D3TPC4 A0A1I8NFB6 A0A0L0BXL2 T1J8D4 E9FU50 Q16WA6 A0A2K7P6C0 A0A1I8QAR4 A0A1B0G3E5 A0A1D2NCD2 A0A1L8DGL6 A0A0P6IHX3 A0A087TNZ7 A0A182H1F3 A0A2I0TWQ1 G1NBF2 A0A226MWM2
A0A212EUZ5 A0A067QNZ6 A0A2J7Q9P9 A0A3L8DBP0 A0A195FIC7 F4W693 A0A026WFV2 A0A151WHS8 A0A195AVL0 A0A158P214 A0A2A3E1S2 A0A088AW37 E9ISC1 A0A1B6EEL2 A0A0J7L6Z8 E2BIM6 A0A1B6BXL4 A0A1B6GLU8 A0A0C9R7A9 K7J1J2 A0A232FIT2 V5GVE7 A0A1E1X2W4 G3MND4 A0A023GK14 A0A131XH63 L7M8T0 A0A224YLY6 A0A131YSF0 A0A1Z5L6P2 A0A293MCY9 A0A1B0CM02 A0A1B0D1L0 A0A2R5LJW0 A0A0A9W5J4 A0A139WEM6 A0A1W4WBN3 A0A1Y1L955 E0VBS2 A0A0P4W2R7 A0A023F4J3 A0A224XPF8 A0A2R7VRS5 A0A069DU68 A0A182PA03 A0A182M913 B0VZR4 A0A0V0G7N5 A0A182WIY4 A0A182QEX3 A0A1Q3FCE4 A0A182RB20 A0A336M572 A0A182XZC6 A0A182UTN6 A0A336MQF3 A0A1C7CZJ7 A0A182UCF7 A0A182X6K8 A0A182I3V7 Q7QJW7 A0A182NHC8 U5EVR2 A0A182JCE3 A0A084VF65 A0A182JTV8 A0A2M4CTH4 A0A336MN00 A0A0K8WG81 A0A182FG29 A0A1B6EA97 A0A1A9W0M8 A0A0P5ZAU3 A0A034W9A0 A0A0A1WL03 A0A0N0BBL1 W8C415 D3TPC4 A0A1I8NFB6 A0A0L0BXL2 T1J8D4 E9FU50 Q16WA6 A0A2K7P6C0 A0A1I8QAR4 A0A1B0G3E5 A0A1D2NCD2 A0A1L8DGL6 A0A0P6IHX3 A0A087TNZ7 A0A182H1F3 A0A2I0TWQ1 G1NBF2 A0A226MWM2
PDB
4OI2
E-value=5.76489e-118,
Score=1086
Ontologies
KEGG
GO
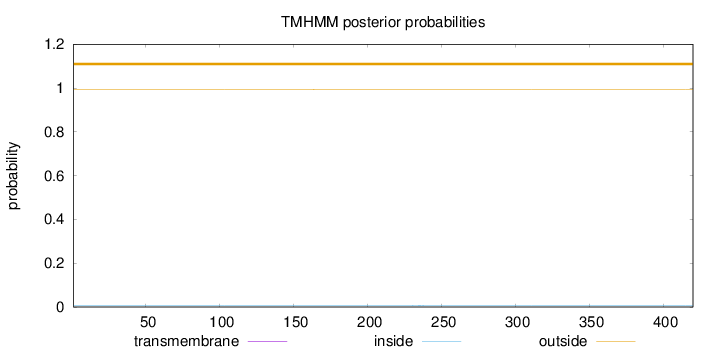
Topology
Subcellular location
Nucleus
Length:
420
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00403999999999999
Exp number, first 60 AAs:
7e-05
Total prob of N-in:
0.00640
outside
1 - 420
Population Genetic Test Statistics
Pi
275.346531
Theta
168.125978
Tajima's D
1.83429
CLR
0.406134
CSRT
0.856057197140143
Interpretation
Uncertain
