Gene
KWMTBOMO14984 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA004782
Annotation
26S_protease_regulatory_subunit_6A_[Danaus_plexippus]
Location in the cell
Cytoplasmic Reliability : 4.222
Sequence
CDS
ATGGCCACAACATTAGAAGACAAGTCAATTTGGGAGGATGGCGAGGAAGCTCTCAGTGAGGAAGTTCTTCGCATGCCAACTGATGAAATCGTAAGTCGAACCCGACTTTTGGACAATGAAATCAAAATAATGAAGAGCGAAGTGATGAGGATCTCGCACGAGCTGCAGGCTCAAAACGATAAAATTAAAGAAAACACGGAGAAAATCAAGGTCAATAAGACGCTACCATATCTCGTATCGAACGTCATTGAACTTTTAGATGTGGATCCGCAAGAGGAGGAAGAAGACGGCGCCGTTGTCGATCTAGACTCACAGCGTAAAGGGAAATGCGCTGTAATCAAGACTTCAACCCGGCAAACATATTTCTTGCCCGTCATCGGCTTGGTAGATGCAGAGAAACTCAAACCAGGAGACTTGGTAGGAGTCAACAAAGACTCTTATTTGATCTTAGAAACGCTCCCAGCGGAGTATGACGCCAGAGTAAAGGCAATGGAGGTAGATGAGAGACCAACTGAACAATACTCAGATATTGGAGGTCTGGATAAACAGATACAGGAATTAATTGAAGCTGTAGTGTTGCCAATGACACACAAGGAGAAATTTGTGAATCTCGGCATCCACCCACCCAAAGGAGTTCTTTTATATGGACCTCCTGGTACAGGGAAGACATTGCTGGCGAGAGCTTGCGCTGCTCAAACAAAGTCTACATTTTTGAAGCTGGCTGGTCCACAACTTGTGCAGATGTTCATTGGTGATGGTGCCAAGCTTGTCCGCGACGCCTTTGCATTGGCTAAAGAGAAGGCACCAGCAATCATCTTCATTGATGAACTGGACGCTATTGGTACAAAGCGTTTTGATTCAGAAAAGGCTGGAGACCGTGAGGTGCAGCGTACTATGTTGGAGTTGTTAAACCAGCTTGATGGGTTTAGCTCAACTGCTGACATCAAGGTCATTGCAGCTACCAATAGAGTGGACATTTTGGACCCTGCCCTGCTCAGATCTGGACGTTTGGACCGAAAAATCGAATTTCCTCATCCCAATGAAGAAGCCCGAGCCAGAATTATGCAGATCCATTCCCGTAAAATGAATGTAAGTCCAGACGTTAATTTCGAAGAACTGTCGCGTTCGACTGATGATTTCAATGGCGCCCAGTGCAAGGCGGTCTGCGTCGAAGCTGGTATGATAGCGCTGCGACGTTCAGCAACTGCCGTCACTCACGAAGACTTTATGGACGCCATACTGGAGGTCCAGGCTAAGAAAAAGGCCAACCTTAGCTATTATGCTTAG
Protein
MATTLEDKSIWEDGEEALSEEVLRMPTDEIVSRTRLLDNEIKIMKSEVMRISHELQAQNDKIKENTEKIKVNKTLPYLVSNVIELLDVDPQEEEEDGAVVDLDSQRKGKCAVIKTSTRQTYFLPVIGLVDAEKLKPGDLVGVNKDSYLILETLPAEYDARVKAMEVDERPTEQYSDIGGLDKQIQELIEAVVLPMTHKEKFVNLGIHPPKGVLLYGPPGTGKTLLARACAAQTKSTFLKLAGPQLVQMFIGDGAKLVRDAFALAKEKAPAIIFIDELDAIGTKRFDSEKAGDREVQRTMLELLNQLDGFSSTADIKVIAATNRVDILDPALLRSGRLDRKIEFPHPNEEARARIMQIHSRKMNVSPDVNFEELSRSTDDFNGAQCKAVCVEAGMIALRRSATAVTHEDFMDAILEVQAKKKANLSYYA
Summary
Similarity
Belongs to the AAA ATPase family.
Uniprot
H9J5J2
A0A212EUZ8
A0A2W1BQ59
A0A2A4JTV8
A0A2H1WL04
S4P9W9
+ More
E3UKM6 A0A194PLU9 A0A023ET77 A0A1L8DSV8 A0A1B0D2F1 A0A1Q3FEI8 B0XA45 A0A1B6C0F7 A0A1W4X464 A0A1B0CC37 Q17HZ7 A0A2J7R3B9 Q299D0 B4G515 B3LY55 B4NG48 B4JT92 B4K5A8 B4M0F9 A0A0M3QYJ8 A0A3B0JL95 B3P8E7 W8BV30 B4R242 U5EUR1 B4PNX2 Q9V3V6 A0A0K8U489 A0A034WDM9 B4HFY0 A0A1I9WLL4 A0A1W4UJE3 A0A232F7V9 A0A0B4J2S7 A0A336JYV2 A0A1Y1LPA0 A0A195D0Z9 E9IJK1 D6WS98 A0A0J7KJ32 A0A158P1E9 E1ZWQ3 A0A026X2J4 A0A182RPF7 A0A0M8ZWQ0 A0A182MZN6 A0A182R091 A0A2A3EML2 A0A088AC90 A0A182W4F3 A0A182M9H7 A0A182SQJ0 A0A182Y7P2 V5GS03 A0A0L0CM06 T1PC96 A0A1I8NWN8 A0A1S3DRG9 A0A182PT41 A0A084VLI2 A0A154P0T0 T1IWS3 A0A182JG51 A0A182KBG6 A0A0K8TMS2 A0A0C9RVS4 A0A0L7QR27 Q9XZC3 A0A182VF47 A0A182WW64 Q7QFQ3 A0A182IA26 E9FRW4 A0A0P5D9G6 A0A310SGF4 A0A1A9XKT2 A0A1A9VQ17 A0A1B0AVB1 A0A0T6AZ01 A0A1A9ZMB1 A0A1A9X2D6 D3TMG4 A0A1J1HUX1 A0A146KVF3 A0A0N8B513 A0A0A9XHW9 A0A0P6FDQ9 A0A0P5SB41 A0A0P5D1Y4 A0A0P5IH79 A0A2M4BNZ5 R4WDQ0 A0A182JG60 A0A0P6C8Q6
E3UKM6 A0A194PLU9 A0A023ET77 A0A1L8DSV8 A0A1B0D2F1 A0A1Q3FEI8 B0XA45 A0A1B6C0F7 A0A1W4X464 A0A1B0CC37 Q17HZ7 A0A2J7R3B9 Q299D0 B4G515 B3LY55 B4NG48 B4JT92 B4K5A8 B4M0F9 A0A0M3QYJ8 A0A3B0JL95 B3P8E7 W8BV30 B4R242 U5EUR1 B4PNX2 Q9V3V6 A0A0K8U489 A0A034WDM9 B4HFY0 A0A1I9WLL4 A0A1W4UJE3 A0A232F7V9 A0A0B4J2S7 A0A336JYV2 A0A1Y1LPA0 A0A195D0Z9 E9IJK1 D6WS98 A0A0J7KJ32 A0A158P1E9 E1ZWQ3 A0A026X2J4 A0A182RPF7 A0A0M8ZWQ0 A0A182MZN6 A0A182R091 A0A2A3EML2 A0A088AC90 A0A182W4F3 A0A182M9H7 A0A182SQJ0 A0A182Y7P2 V5GS03 A0A0L0CM06 T1PC96 A0A1I8NWN8 A0A1S3DRG9 A0A182PT41 A0A084VLI2 A0A154P0T0 T1IWS3 A0A182JG51 A0A182KBG6 A0A0K8TMS2 A0A0C9RVS4 A0A0L7QR27 Q9XZC3 A0A182VF47 A0A182WW64 Q7QFQ3 A0A182IA26 E9FRW4 A0A0P5D9G6 A0A310SGF4 A0A1A9XKT2 A0A1A9VQ17 A0A1B0AVB1 A0A0T6AZ01 A0A1A9ZMB1 A0A1A9X2D6 D3TMG4 A0A1J1HUX1 A0A146KVF3 A0A0N8B513 A0A0A9XHW9 A0A0P6FDQ9 A0A0P5SB41 A0A0P5D1Y4 A0A0P5IH79 A0A2M4BNZ5 R4WDQ0 A0A182JG60 A0A0P6C8Q6
Pubmed
19121390
22118469
28756777
23622113
26354079
24945155
+ More
26483478 17510324 15632085 17994087 18057021 24495485 17550304 10893261 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25348373 27538518 28648823 20075255 28004739 21282665 18362917 19820115 21347285 20798317 24508170 30249741 25244985 26108605 25315136 24438588 26369729 12364791 14747013 17210077 21292972 20353571 26823975 25401762 23691247
26483478 17510324 15632085 17994087 18057021 24495485 17550304 10893261 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25348373 27538518 28648823 20075255 28004739 21282665 18362917 19820115 21347285 20798317 24508170 30249741 25244985 26108605 25315136 24438588 26369729 12364791 14747013 17210077 21292972 20353571 26823975 25401762 23691247
EMBL
BABH01025250
AGBW02012265
OWR45309.1
KZ150023
PZC74860.1
NWSH01000636
+ More
PCG75128.1 ODYU01009335 SOQ53687.1 GAIX01003524 JAA89036.1 HM449900 ADO33035.1 KQ459604 KPI92080.1 JXUM01068691 GAPW01001577 KQ562513 JAC12021.1 KXJ75734.1 GFDF01004541 JAV09543.1 AJVK01022777 GFDL01009081 JAV25964.1 DS232564 EDS43510.1 GEDC01030444 GEDC01029000 JAS06854.1 JAS08298.1 AJWK01006101 CH477244 EAT46279.1 NEVH01007821 PNF35338.1 CM000070 EAL27773.1 CH479179 EDW24681.1 CH902617 EDV43959.1 CH964251 EDW83265.1 CH916373 EDV94982.1 CH933806 EDW16134.1 KRG01952.1 CH940650 EDW68338.1 KRF83786.1 CP012526 ALC47803.1 OUUW01000007 SPP83037.1 CH954182 EDV53971.1 GAMC01001350 JAC05206.1 CM000364 EDX14099.1 GANO01002208 JAB57663.1 CM000160 EDW98182.1 AF160882 AF145305 AE014297 AAD46823.1 AAF08386.1 AAF56177.1 GDHF01030936 JAI21378.1 GAKP01005281 JAC53671.1 CH480815 EDW43373.1 KU932398 APA34034.1 NNAY01000782 OXU26539.1 UFQS01000014 UFQT01000014 SSW97297.1 SSX17683.1 GEZM01051891 JAV74648.1 KQ976986 KYN06583.1 GL763868 EFZ19244.1 KQ971354 EFA07110.1 LBMM01006642 KMQ90438.1 ADTU01006397 GL434886 EFN74381.1 KK107024 QOIP01000004 EZA62221.1 RLU23390.1 KQ435834 KOX71606.1 AXCN02001328 KZ288210 PBC32968.1 AXCM01002197 GALX01005453 JAB63013.1 JRES01000196 KNC33368.1 KA645750 AFP60379.1 ATLV01014499 KE524974 KFB38826.1 KQ434796 KZC05535.1 JH431629 GDAI01002157 JAI15446.1 GBYB01011806 JAG81573.1 KQ414784 KOC61082.1 AF134402 AAD24194.1 AAAB01008846 EAA06390.3 APCN01001092 GL732523 EFX89917.1 GDIP01165046 GDIP01159495 GDIP01159120 JAJ64282.1 KQ764369 OAD54530.1 JXJN01004168 LJIG01022515 KRT80171.1 CCAG010021212 EZ422616 ADD18892.1 CVRI01000021 CRK91871.1 GDHC01019413 JAP99215.1 GDIQ01212155 GDIQ01036878 JAK39570.1 GBHO01026934 JAG16670.1 GDIQ01159631 GDIQ01061370 LRGB01001654 JAN33367.1 KZS10945.1 GDIQ01106695 JAL45031.1 GDIP01166587 JAJ56815.1 GDIQ01214356 JAK37369.1 GGFJ01005591 MBW54732.1 AK417734 BAN20949.1 GDIP01005571 JAM98144.1
PCG75128.1 ODYU01009335 SOQ53687.1 GAIX01003524 JAA89036.1 HM449900 ADO33035.1 KQ459604 KPI92080.1 JXUM01068691 GAPW01001577 KQ562513 JAC12021.1 KXJ75734.1 GFDF01004541 JAV09543.1 AJVK01022777 GFDL01009081 JAV25964.1 DS232564 EDS43510.1 GEDC01030444 GEDC01029000 JAS06854.1 JAS08298.1 AJWK01006101 CH477244 EAT46279.1 NEVH01007821 PNF35338.1 CM000070 EAL27773.1 CH479179 EDW24681.1 CH902617 EDV43959.1 CH964251 EDW83265.1 CH916373 EDV94982.1 CH933806 EDW16134.1 KRG01952.1 CH940650 EDW68338.1 KRF83786.1 CP012526 ALC47803.1 OUUW01000007 SPP83037.1 CH954182 EDV53971.1 GAMC01001350 JAC05206.1 CM000364 EDX14099.1 GANO01002208 JAB57663.1 CM000160 EDW98182.1 AF160882 AF145305 AE014297 AAD46823.1 AAF08386.1 AAF56177.1 GDHF01030936 JAI21378.1 GAKP01005281 JAC53671.1 CH480815 EDW43373.1 KU932398 APA34034.1 NNAY01000782 OXU26539.1 UFQS01000014 UFQT01000014 SSW97297.1 SSX17683.1 GEZM01051891 JAV74648.1 KQ976986 KYN06583.1 GL763868 EFZ19244.1 KQ971354 EFA07110.1 LBMM01006642 KMQ90438.1 ADTU01006397 GL434886 EFN74381.1 KK107024 QOIP01000004 EZA62221.1 RLU23390.1 KQ435834 KOX71606.1 AXCN02001328 KZ288210 PBC32968.1 AXCM01002197 GALX01005453 JAB63013.1 JRES01000196 KNC33368.1 KA645750 AFP60379.1 ATLV01014499 KE524974 KFB38826.1 KQ434796 KZC05535.1 JH431629 GDAI01002157 JAI15446.1 GBYB01011806 JAG81573.1 KQ414784 KOC61082.1 AF134402 AAD24194.1 AAAB01008846 EAA06390.3 APCN01001092 GL732523 EFX89917.1 GDIP01165046 GDIP01159495 GDIP01159120 JAJ64282.1 KQ764369 OAD54530.1 JXJN01004168 LJIG01022515 KRT80171.1 CCAG010021212 EZ422616 ADD18892.1 CVRI01000021 CRK91871.1 GDHC01019413 JAP99215.1 GDIQ01212155 GDIQ01036878 JAK39570.1 GBHO01026934 JAG16670.1 GDIQ01159631 GDIQ01061370 LRGB01001654 JAN33367.1 KZS10945.1 GDIQ01106695 JAL45031.1 GDIP01166587 JAJ56815.1 GDIQ01214356 JAK37369.1 GGFJ01005591 MBW54732.1 AK417734 BAN20949.1 GDIP01005571 JAM98144.1
Proteomes
UP000005204
UP000007151
UP000218220
UP000053268
UP000069940
UP000249989
+ More
UP000092462 UP000002320 UP000192223 UP000092461 UP000008820 UP000235965 UP000001819 UP000008744 UP000007801 UP000007798 UP000001070 UP000009192 UP000008792 UP000092553 UP000268350 UP000008711 UP000000304 UP000002282 UP000000803 UP000001292 UP000192221 UP000215335 UP000002358 UP000078542 UP000007266 UP000036403 UP000005205 UP000000311 UP000053097 UP000279307 UP000075900 UP000053105 UP000075884 UP000075886 UP000242457 UP000005203 UP000075920 UP000075883 UP000075901 UP000076408 UP000037069 UP000095301 UP000095300 UP000079169 UP000075885 UP000030765 UP000076502 UP000075880 UP000075881 UP000053825 UP000075903 UP000076407 UP000007062 UP000075840 UP000000305 UP000092443 UP000078200 UP000092460 UP000092445 UP000091820 UP000092444 UP000183832 UP000076858
UP000092462 UP000002320 UP000192223 UP000092461 UP000008820 UP000235965 UP000001819 UP000008744 UP000007801 UP000007798 UP000001070 UP000009192 UP000008792 UP000092553 UP000268350 UP000008711 UP000000304 UP000002282 UP000000803 UP000001292 UP000192221 UP000215335 UP000002358 UP000078542 UP000007266 UP000036403 UP000005205 UP000000311 UP000053097 UP000279307 UP000075900 UP000053105 UP000075884 UP000075886 UP000242457 UP000005203 UP000075920 UP000075883 UP000075901 UP000076408 UP000037069 UP000095301 UP000095300 UP000079169 UP000075885 UP000030765 UP000076502 UP000075880 UP000075881 UP000053825 UP000075903 UP000076407 UP000007062 UP000075840 UP000000305 UP000092443 UP000078200 UP000092460 UP000092445 UP000091820 UP000092444 UP000183832 UP000076858
PRIDE
Interpro
SUPFAM
SSF52540
SSF52540
ProteinModelPortal
H9J5J2
A0A212EUZ8
A0A2W1BQ59
A0A2A4JTV8
A0A2H1WL04
S4P9W9
+ More
E3UKM6 A0A194PLU9 A0A023ET77 A0A1L8DSV8 A0A1B0D2F1 A0A1Q3FEI8 B0XA45 A0A1B6C0F7 A0A1W4X464 A0A1B0CC37 Q17HZ7 A0A2J7R3B9 Q299D0 B4G515 B3LY55 B4NG48 B4JT92 B4K5A8 B4M0F9 A0A0M3QYJ8 A0A3B0JL95 B3P8E7 W8BV30 B4R242 U5EUR1 B4PNX2 Q9V3V6 A0A0K8U489 A0A034WDM9 B4HFY0 A0A1I9WLL4 A0A1W4UJE3 A0A232F7V9 A0A0B4J2S7 A0A336JYV2 A0A1Y1LPA0 A0A195D0Z9 E9IJK1 D6WS98 A0A0J7KJ32 A0A158P1E9 E1ZWQ3 A0A026X2J4 A0A182RPF7 A0A0M8ZWQ0 A0A182MZN6 A0A182R091 A0A2A3EML2 A0A088AC90 A0A182W4F3 A0A182M9H7 A0A182SQJ0 A0A182Y7P2 V5GS03 A0A0L0CM06 T1PC96 A0A1I8NWN8 A0A1S3DRG9 A0A182PT41 A0A084VLI2 A0A154P0T0 T1IWS3 A0A182JG51 A0A182KBG6 A0A0K8TMS2 A0A0C9RVS4 A0A0L7QR27 Q9XZC3 A0A182VF47 A0A182WW64 Q7QFQ3 A0A182IA26 E9FRW4 A0A0P5D9G6 A0A310SGF4 A0A1A9XKT2 A0A1A9VQ17 A0A1B0AVB1 A0A0T6AZ01 A0A1A9ZMB1 A0A1A9X2D6 D3TMG4 A0A1J1HUX1 A0A146KVF3 A0A0N8B513 A0A0A9XHW9 A0A0P6FDQ9 A0A0P5SB41 A0A0P5D1Y4 A0A0P5IH79 A0A2M4BNZ5 R4WDQ0 A0A182JG60 A0A0P6C8Q6
E3UKM6 A0A194PLU9 A0A023ET77 A0A1L8DSV8 A0A1B0D2F1 A0A1Q3FEI8 B0XA45 A0A1B6C0F7 A0A1W4X464 A0A1B0CC37 Q17HZ7 A0A2J7R3B9 Q299D0 B4G515 B3LY55 B4NG48 B4JT92 B4K5A8 B4M0F9 A0A0M3QYJ8 A0A3B0JL95 B3P8E7 W8BV30 B4R242 U5EUR1 B4PNX2 Q9V3V6 A0A0K8U489 A0A034WDM9 B4HFY0 A0A1I9WLL4 A0A1W4UJE3 A0A232F7V9 A0A0B4J2S7 A0A336JYV2 A0A1Y1LPA0 A0A195D0Z9 E9IJK1 D6WS98 A0A0J7KJ32 A0A158P1E9 E1ZWQ3 A0A026X2J4 A0A182RPF7 A0A0M8ZWQ0 A0A182MZN6 A0A182R091 A0A2A3EML2 A0A088AC90 A0A182W4F3 A0A182M9H7 A0A182SQJ0 A0A182Y7P2 V5GS03 A0A0L0CM06 T1PC96 A0A1I8NWN8 A0A1S3DRG9 A0A182PT41 A0A084VLI2 A0A154P0T0 T1IWS3 A0A182JG51 A0A182KBG6 A0A0K8TMS2 A0A0C9RVS4 A0A0L7QR27 Q9XZC3 A0A182VF47 A0A182WW64 Q7QFQ3 A0A182IA26 E9FRW4 A0A0P5D9G6 A0A310SGF4 A0A1A9XKT2 A0A1A9VQ17 A0A1B0AVB1 A0A0T6AZ01 A0A1A9ZMB1 A0A1A9X2D6 D3TMG4 A0A1J1HUX1 A0A146KVF3 A0A0N8B513 A0A0A9XHW9 A0A0P6FDQ9 A0A0P5SB41 A0A0P5D1Y4 A0A0P5IH79 A0A2M4BNZ5 R4WDQ0 A0A182JG60 A0A0P6C8Q6
PDB
6MSK
E-value=0,
Score=1834
Ontologies
GO
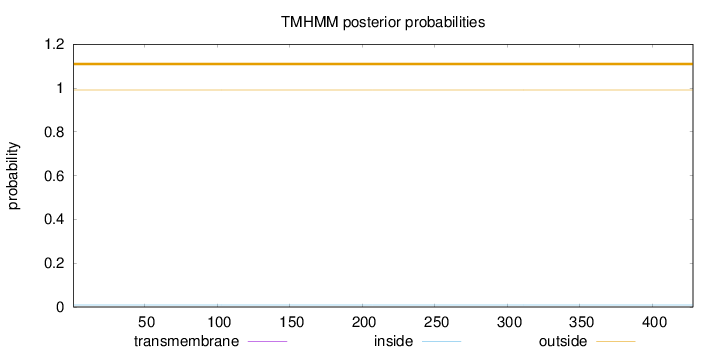
Topology
Length:
428
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00132
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00946
outside
1 - 428
Population Genetic Test Statistics
Pi
322.940808
Theta
234.670365
Tajima's D
0.994332
CLR
0.320925
CSRT
0.662116894155292
Interpretation
Uncertain
