Pre Gene Modal
BGIBMGA004884
Annotation
hypothetical_protein_KGM_22037_[Danaus_plexippus]
Full name
Succinate dehydrogenase [ubiquinone] cytochrome b small subunit
Location in the cell
PlasmaMembrane Reliability : 4.13
Sequence
CDS
ATGGCGTTCTCCATGTTTCTTCGTACATCAGGATGCCAAAGTCGAATGTTCACTCAACAAGCTATGAGGCTAGCGACCCAGCCAGCAATAACACAACGTATGATCTCCAGCTCATTGCCTGTTAGAAATATAATGAAGGATACTAAAACGACACCCATATTGAATGCTATTCGTTCATTCCGTACGTCTTGCGTGCGCCGGTCAGCGGAGAAGGTCGAGGACCACTCCAGGCTCTGGGTCATTGAGAGAGTGGTTTCTGCTGCACTGGTCCCATTGATTCCGTTAGCCCTCATGATGCCGAACAAACTATTCGACTCTCTCCTGGCGATACTGATCACCGCTCACAGTTTCTGGGGCCTGGAAGCGATCGCGGTCGACTACGTGCGCGCTAGTATATTTGGTCCTATTTTACCCAAAATTGCCATAGGCCTGGTGTATCTCATCTCCATTGCCACACTGGGAGGCTTGTTTTACATCATCAGCCACGATGTCGGCATCGCCAACGGCATCAGACAGTTCTGGGCCATCAAATCGGACGCACAGAAGTCATAA
Protein
MAFSMFLRTSGCQSRMFTQQAMRLATQPAITQRMISSSLPVRNIMKDTKTTPILNAIRSFRTSCVRRSAEKVEDHSRLWVIERVVSAALVPLIPLALMMPNKLFDSLLAILITAHSFWGLEAIAVDYVRASIFGPILPKIAIGLVYLISIATLGGLFYIISHDVGIANGIRQFWAIKSDAQKS
Summary
Description
Membrane-anchoring subunit of succinate dehydrogenase (SDH) that is involved in complex II of the mitochondrial electron transport chain and is responsible for transferring electrons from succinate to ubiquinone (coenzyme Q).
Similarity
Belongs to the CybS family.
Feature
chain Succinate dehydrogenase [ubiquinone] cytochrome b small subunit
Uniprot
A0A212EUX9
A0A3S2M986
A0A194RI39
L0CRD8
S4P5H2
A0A1E1WR06
+ More
A0A194PFD8 A0A2A4JSV9 A0A2W1BKQ6 A0A0L7KN38 A0A2H1WL42 A0A212FDM2 A0A034VUA1 A0A0A1XHK1 B3P8E4 A0A0K8WKU5 B4PNX5 Q1HR55 Q16SP7 A0A182H1A0 A0A067R7U5 A0A023EI85 A0A226EIG9 A0A0P4VVR8 A0A023EI91 A0A1I8QAZ5 A0A3B0JRP6 Q6XHN2 A0A023EGE8 D6WTA5 A0A0Q9WIL1 A0A1Q3FG07 B4M0F6 A0A1Q3FFN0 A0A1B6D8P8 E9GWY0 A0A1A9W2N4 A0A0K8TPB5 A0A2R7W2E2 A0A182PT43 A0A1J1HJY9 A0A2M4AHU1 A0A182Y7P4 A0A182K2H4 E0VV75 A0A170YAQ0 A0A182FIX8 A0A182WW66 A0A182W4F5 A0A182M7F2 A0A293LCA3 A0A182RPF9 A0A2R5LHM8 A0A1Y3BFJ2
A0A194PFD8 A0A2A4JSV9 A0A2W1BKQ6 A0A0L7KN38 A0A2H1WL42 A0A212FDM2 A0A034VUA1 A0A0A1XHK1 B3P8E4 A0A0K8WKU5 B4PNX5 Q1HR55 Q16SP7 A0A182H1A0 A0A067R7U5 A0A023EI85 A0A226EIG9 A0A0P4VVR8 A0A023EI91 A0A1I8QAZ5 A0A3B0JRP6 Q6XHN2 A0A023EGE8 D6WTA5 A0A0Q9WIL1 A0A1Q3FG07 B4M0F6 A0A1Q3FFN0 A0A1B6D8P8 E9GWY0 A0A1A9W2N4 A0A0K8TPB5 A0A2R7W2E2 A0A182PT43 A0A1J1HJY9 A0A2M4AHU1 A0A182Y7P4 A0A182K2H4 E0VV75 A0A170YAQ0 A0A182FIX8 A0A182WW66 A0A182W4F5 A0A182M7F2 A0A293LCA3 A0A182RPF9 A0A2R5LHM8 A0A1Y3BFJ2
Pubmed
EMBL
AGBW02012265
OWR45308.1
RSAL01000009
RVE53855.1
KQ460205
KPJ16995.1
+ More
JX870003 JX870004 JX870005 JX870006 JX870007 AGA19582.1 GAIX01008622 JAA83938.1 GDQN01001614 JAT89440.1 KQ459604 KPI92081.1 NWSH01000636 PCG75127.1 KZ150023 PZC74861.1 JTDY01008186 KOB64692.1 ODYU01009335 SOQ53686.1 AGBW02009011 OWR51875.1 GAKP01012046 GAKP01012044 JAC46906.1 GBXI01003855 JAD10437.1 CH954182 EDV53968.1 GDHF01000635 JAI51679.1 CM000160 EDW98185.1 DQ440239 CH478321 ABF18272.1 EAT33234.1 CH477669 EAT37500.1 JXUM01037208 JXUM01102818 KQ564754 KQ561121 KXJ71783.1 KXJ79642.1 KK852683 KDR18542.1 GAPW01004968 JAC08630.1 LNIX01000004 OXA56346.1 GDKW01000446 JAI56149.1 GAPW01004967 JAC08631.1 OUUW01000007 SPP83042.1 AY232151 AAR10174.1 GAPW01005171 JAC08427.1 KQ971352 EFA07507.1 CH940650 KRF83782.1 GFDL01008561 JAV26484.1 EDW68335.1 GFDL01008692 JAV26353.1 GEDC01015224 JAS22074.1 GL732571 EFX76036.1 GDAI01001617 JAI15986.1 KK854252 PTY13749.1 CVRI01000006 CRK88341.1 GGFK01006996 MBW40317.1 DS235804 EEB17281.1 GEMB01003486 JAR99729.1 AXCM01002197 GFWV01001603 MAA26333.1 GGLE01004880 MBY09006.1 MUJZ01022107 OTF79632.1
JX870003 JX870004 JX870005 JX870006 JX870007 AGA19582.1 GAIX01008622 JAA83938.1 GDQN01001614 JAT89440.1 KQ459604 KPI92081.1 NWSH01000636 PCG75127.1 KZ150023 PZC74861.1 JTDY01008186 KOB64692.1 ODYU01009335 SOQ53686.1 AGBW02009011 OWR51875.1 GAKP01012046 GAKP01012044 JAC46906.1 GBXI01003855 JAD10437.1 CH954182 EDV53968.1 GDHF01000635 JAI51679.1 CM000160 EDW98185.1 DQ440239 CH478321 ABF18272.1 EAT33234.1 CH477669 EAT37500.1 JXUM01037208 JXUM01102818 KQ564754 KQ561121 KXJ71783.1 KXJ79642.1 KK852683 KDR18542.1 GAPW01004968 JAC08630.1 LNIX01000004 OXA56346.1 GDKW01000446 JAI56149.1 GAPW01004967 JAC08631.1 OUUW01000007 SPP83042.1 AY232151 AAR10174.1 GAPW01005171 JAC08427.1 KQ971352 EFA07507.1 CH940650 KRF83782.1 GFDL01008561 JAV26484.1 EDW68335.1 GFDL01008692 JAV26353.1 GEDC01015224 JAS22074.1 GL732571 EFX76036.1 GDAI01001617 JAI15986.1 KK854252 PTY13749.1 CVRI01000006 CRK88341.1 GGFK01006996 MBW40317.1 DS235804 EEB17281.1 GEMB01003486 JAR99729.1 AXCM01002197 GFWV01001603 MAA26333.1 GGLE01004880 MBY09006.1 MUJZ01022107 OTF79632.1
Proteomes
UP000007151
UP000283053
UP000053240
UP000053268
UP000218220
UP000037510
+ More
UP000008711 UP000002282 UP000008820 UP000069940 UP000249989 UP000027135 UP000198287 UP000095300 UP000268350 UP000007266 UP000008792 UP000000305 UP000091820 UP000075885 UP000183832 UP000076408 UP000075881 UP000009046 UP000069272 UP000076407 UP000075920 UP000075883 UP000075900
UP000008711 UP000002282 UP000008820 UP000069940 UP000249989 UP000027135 UP000198287 UP000095300 UP000268350 UP000007266 UP000008792 UP000000305 UP000091820 UP000075885 UP000183832 UP000076408 UP000075881 UP000009046 UP000069272 UP000076407 UP000075920 UP000075883 UP000075900
Pfam
PF00595 PDZ
Interpro
Gene 3D
ProteinModelPortal
A0A212EUX9
A0A3S2M986
A0A194RI39
L0CRD8
S4P5H2
A0A1E1WR06
+ More
A0A194PFD8 A0A2A4JSV9 A0A2W1BKQ6 A0A0L7KN38 A0A2H1WL42 A0A212FDM2 A0A034VUA1 A0A0A1XHK1 B3P8E4 A0A0K8WKU5 B4PNX5 Q1HR55 Q16SP7 A0A182H1A0 A0A067R7U5 A0A023EI85 A0A226EIG9 A0A0P4VVR8 A0A023EI91 A0A1I8QAZ5 A0A3B0JRP6 Q6XHN2 A0A023EGE8 D6WTA5 A0A0Q9WIL1 A0A1Q3FG07 B4M0F6 A0A1Q3FFN0 A0A1B6D8P8 E9GWY0 A0A1A9W2N4 A0A0K8TPB5 A0A2R7W2E2 A0A182PT43 A0A1J1HJY9 A0A2M4AHU1 A0A182Y7P4 A0A182K2H4 E0VV75 A0A170YAQ0 A0A182FIX8 A0A182WW66 A0A182W4F5 A0A182M7F2 A0A293LCA3 A0A182RPF9 A0A2R5LHM8 A0A1Y3BFJ2
A0A194PFD8 A0A2A4JSV9 A0A2W1BKQ6 A0A0L7KN38 A0A2H1WL42 A0A212FDM2 A0A034VUA1 A0A0A1XHK1 B3P8E4 A0A0K8WKU5 B4PNX5 Q1HR55 Q16SP7 A0A182H1A0 A0A067R7U5 A0A023EI85 A0A226EIG9 A0A0P4VVR8 A0A023EI91 A0A1I8QAZ5 A0A3B0JRP6 Q6XHN2 A0A023EGE8 D6WTA5 A0A0Q9WIL1 A0A1Q3FG07 B4M0F6 A0A1Q3FFN0 A0A1B6D8P8 E9GWY0 A0A1A9W2N4 A0A0K8TPB5 A0A2R7W2E2 A0A182PT43 A0A1J1HJY9 A0A2M4AHU1 A0A182Y7P4 A0A182K2H4 E0VV75 A0A170YAQ0 A0A182FIX8 A0A182WW66 A0A182W4F5 A0A182M7F2 A0A293LCA3 A0A182RPF9 A0A2R5LHM8 A0A1Y3BFJ2
PDB
5C3J
E-value=2.47596e-11,
Score=161
Ontologies
PATHWAY
GO
Topology
Subcellular location
Mitochondrion inner membrane
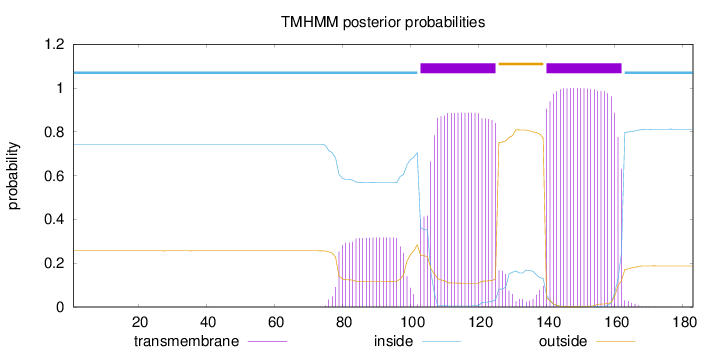
Length:
183
Number of predicted TMHs:
2
Exp number of AAs in TMHs:
48.06255
Exp number, first 60 AAs:
0.00748
Total prob of N-in:
0.74316
inside
1 - 102
TMhelix
103 - 125
outside
126 - 139
TMhelix
140 - 162
inside
163 - 183
Population Genetic Test Statistics
Pi
279.64087
Theta
196.491009
Tajima's D
1.390927
CLR
0.168384
CSRT
0.760861956902155
Interpretation
Uncertain
