Gene
KWMTBOMO14982
Pre Gene Modal
BGIBMGA004781
Annotation
PREDICTED:_phosphatidylglycerophosphatase_and_protein-tyrosine_phosphatase_1_[Amyelois_transitella]
Full name
Phosphatidylglycerophosphatase and protein-tyrosine phosphatase 1
Alternative Name
PTEN-like phosphatase
PTEN-like protein
Protein-tyrosine phosphatase mitochondrial 1-like protein
PTEN-like protein
Protein-tyrosine phosphatase mitochondrial 1-like protein
Location in the cell
Cytoplasmic Reliability : 1.085 Mitochondrial Reliability : 1.555
Sequence
CDS
ATGAGTACGGCAATGTTCGCGAGAGTTACATTCTACCCGACCCTTTTGTACAATGTCTTCATGGAGAAGGTGACCAGTAGACGCTGGTATGACAGAATCGATGACACGGTTATCCTAGGAGCCCTGCCCTTTCAAGGAATGACAAAACAGTTAAAAGAAGAGGAGAATGTAAAGGGAGTTGTCTCAATGAATGAGACATATGAGCTACAATTGTTCTCTAATGATGCAGAGAGATGGCGAGAACACAAAGTGGAATTCCTACAGTTGGCCACAACAGATATTTTTGAGGCCCCCAATCAAGACAAATTATTCGAAGGAGTCGTGTTTATAAACAGATTCCTGCCGTTAGACAACAAACTGTCGGGAGTCCCTGCCGACGTGGGGCAGATCAACACCGGCACCGTATACGTTCACTGCAAGGCGGGTAGGACGAGAAGCGCCACGCTCGTGGGCTGCTACCTTATGATGAAAAACGGTTGGTCACCCCACGAAGCGGTAGAGTACATGAGATCGAGGCGTCCACACATACTTCTCCACACGAAGCAGTGGGAGGCCCTCGATATATTTCACAGACAACATGTGCGGACGTGA
Protein
MSTAMFARVTFYPTLLYNVFMEKVTSRRWYDRIDDTVILGALPFQGMTKQLKEEENVKGVVSMNETYELQLFSNDAERWREHKVEFLQLATTDIFEAPNQDKLFEGVVFINRFLPLDNKLSGVPADVGQINTGTVYVHCKAGRTRSATLVGCYLMMKNGWSPHEAVEYMRSRRPHILLHTKQWEALDIFHRQHVRT
Summary
Description
Lipid phosphatase that may mediate dephosphorylation of mitochondrial proteins (By similarity). Protein phosphatase that may mediate dephosphorylation of mitochondrial proteins (By similarity). Does not dephosphorylate Akt.
Catalytic Activity
1,2-diacyl-sn-glycero-3-phospho-(1'-sn-glycero-3'-phosphate) + H2O = 1,2-diacyl-sn-glycero-3-phospho-(1'-sn-glycerol) + phosphate
H2O + O-phospho-L-tyrosyl-[protein] = L-tyrosyl-[protein] + phosphate
H2O + O-phospho-L-seryl-[protein] = L-seryl-[protein] + phosphate
H2O + O-phospho-L-threonyl-[protein] = L-threonyl-[protein] + phosphate
H2O + O-phospho-L-tyrosyl-[protein] = L-tyrosyl-[protein] + phosphate
H2O + O-phospho-L-seryl-[protein] = L-seryl-[protein] + phosphate
H2O + O-phospho-L-threonyl-[protein] = L-threonyl-[protein] + phosphate
Similarity
Belongs to the protein-tyrosine phosphatase family. Non-receptor class dual specificity subfamily.
Keywords
Alternative splicing
Complete proteome
Hydrolase
Lipid biosynthesis
Lipid metabolism
Membrane
Mitochondrion
Mitochondrion inner membrane
Phospholipid biosynthesis
Phospholipid metabolism
Protein phosphatase
Reference proteome
Transit peptide
Feature
chain Phosphatidylglycerophosphatase and protein-tyrosine phosphatase 1
splice variant In isoform A.
splice variant In isoform A.
Uniprot
A0A2A4JSU5
A0A2A4JTL5
A0A3S2TSF3
A0A212EV24
H9J5J1
A0A034VWS7
+ More
B4G513 A0A034VXS0 Q299C8 A0A0K8WF45 A0A3B0KAC6 A0A3B0K4W4 U5EYC1 A0A0A1XNP6 A0A0K8U8J9 A0A3B0KDF1 A0A1I8P7H9 A0A0A1WZ67 B4PNX4 B5RJ36 Q86BN8 B3LY53 B4R244 B3P8E5 A0A1I8M2Q8 A0A0L0BUN9 B4HFY2 B4M0F7 A0A336LYB7 Q86BN8-2 A0A0K8TTQ9 B4K5B0 A0A1W4U878 B4JT94 B4NG46 W8BZG4 A0A0Q9WP77 A0A1Q3FJ38 A0A2M3ZCP1 A0A2M4C097 A0A2M4C0B0 T1EB78 A0A182M0E1 A0A1L8E3E9 A0A1W4UL73 E2B7L8 F5HJE0 A0A182FY75 E2AK13 A0A2M4ATL7 A0A1W7R8S0 A0A182RPF8 A0A182UV99 A0A1A9VM33 Q7PRY8 D3TQZ2 A0A2M4CVI2 A0A151WXU2 A0A182PT42 A0A2M4CVH1 A0A2H1WVW9 Q16G69 Q16SP8 A0A1A9Y7H3 B0W9E6 A0A182K2H5 A0A182QGP3 A0A182Y7P3 A0A182IA25 A0A182WW65 A0A1B0CC35 V9ICB3 A0A1B0C7X0 A0A182W4F4 A0A182TTT3 A0A195F850 K7IR65 A0A1S4FQZ6 E9IM00 A0A310SGT1 A0A154PPI4 A0A1B6MG75 A0A2A3E741 A0A194PFM1 A0A195CIE6 A0A158NP97 A0A088A7J8 A0A195AY43 E0VV74 A0A1B6I4T3 A0A0A9W6U2 A0A182MZN7 A0A1B6GP30 A0A023F738 A0A069DWV9 A0A146MC61 A0A0P4VU03 A0A232F132 A0A0M9A041 A0A026VUG6
B4G513 A0A034VXS0 Q299C8 A0A0K8WF45 A0A3B0KAC6 A0A3B0K4W4 U5EYC1 A0A0A1XNP6 A0A0K8U8J9 A0A3B0KDF1 A0A1I8P7H9 A0A0A1WZ67 B4PNX4 B5RJ36 Q86BN8 B3LY53 B4R244 B3P8E5 A0A1I8M2Q8 A0A0L0BUN9 B4HFY2 B4M0F7 A0A336LYB7 Q86BN8-2 A0A0K8TTQ9 B4K5B0 A0A1W4U878 B4JT94 B4NG46 W8BZG4 A0A0Q9WP77 A0A1Q3FJ38 A0A2M3ZCP1 A0A2M4C097 A0A2M4C0B0 T1EB78 A0A182M0E1 A0A1L8E3E9 A0A1W4UL73 E2B7L8 F5HJE0 A0A182FY75 E2AK13 A0A2M4ATL7 A0A1W7R8S0 A0A182RPF8 A0A182UV99 A0A1A9VM33 Q7PRY8 D3TQZ2 A0A2M4CVI2 A0A151WXU2 A0A182PT42 A0A2M4CVH1 A0A2H1WVW9 Q16G69 Q16SP8 A0A1A9Y7H3 B0W9E6 A0A182K2H5 A0A182QGP3 A0A182Y7P3 A0A182IA25 A0A182WW65 A0A1B0CC35 V9ICB3 A0A1B0C7X0 A0A182W4F4 A0A182TTT3 A0A195F850 K7IR65 A0A1S4FQZ6 E9IM00 A0A310SGT1 A0A154PPI4 A0A1B6MG75 A0A2A3E741 A0A194PFM1 A0A195CIE6 A0A158NP97 A0A088A7J8 A0A195AY43 E0VV74 A0A1B6I4T3 A0A0A9W6U2 A0A182MZN7 A0A1B6GP30 A0A023F738 A0A069DWV9 A0A146MC61 A0A0P4VU03 A0A232F132 A0A0M9A041 A0A026VUG6
EC Number
3.1.3.27
Pubmed
22118469
19121390
25348373
17994087
15632085
25830018
+ More
17550304 10731132 12537572 12537569 15247229 25315136 26108605 26369729 24495485 20798317 12364791 14747013 17210077 20353571 17510324 25244985 20075255 21282665 26354079 21347285 20566863 25401762 26823975 25474469 26334808 27129103 28648823 24508170
17550304 10731132 12537572 12537569 15247229 25315136 26108605 26369729 24495485 20798317 12364791 14747013 17210077 20353571 17510324 25244985 20075255 21282665 26354079 21347285 20566863 25401762 26823975 25474469 26334808 27129103 28648823 24508170
EMBL
NWSH01000636
PCG75115.1
PCG75116.1
RSAL01000009
RVE53854.1
AGBW02012265
+ More
OWR45307.1 BABH01025250 BABH01025251 BABH01025252 GAKP01012043 JAC46909.1 CH479179 EDW24679.1 GAKP01012045 JAC46907.1 CM000070 EAL27775.2 GDHF01002640 JAI49674.1 OUUW01000007 SPP83039.1 SPP83040.1 GANO01002093 JAB57778.1 GBXI01001720 JAD12572.1 GDHF01029322 JAI22992.1 SPP83041.1 GBXI01011741 GBXI01010512 GBXI01002214 JAD02551.1 JAD03780.1 JAD12078.1 CM000160 EDW98184.1 BT044310 ACH92375.1 AE014297 AY071042 CH902617 EDV43957.1 CM000364 EDX14101.1 CH954182 EDV53969.1 JRES01001300 KNC23785.1 CH480815 EDW43375.1 CH940650 EDW68336.1 UFQS01000205 UFQT01000205 SSX01329.1 SSX21709.1 GDAI01000070 JAI17533.1 CH933806 EDW16136.1 CH916373 EDV94984.1 CH964251 EDW83263.1 GAMC01001818 JAC04738.1 KRF83783.1 GFDL01007549 JAV27496.1 GGFM01005521 MBW26272.1 GGFJ01009608 MBW58749.1 GGFJ01009591 MBW58732.1 GAMD01000173 JAB01418.1 AXCM01002197 GFDF01000982 JAV13102.1 GL446193 EFN88296.1 AAAB01008846 EGK96401.1 GL440135 EFN66223.1 GGFK01010809 MBW44130.1 GEHC01000111 JAV47534.1 EAA06392.5 CCAG010012076 EZ423844 ADD20120.1 GGFL01005101 MBW69279.1 KQ982668 KYQ52511.1 GGFL01005023 MBW69201.1 ODYU01011413 SOQ57142.1 CH478321 EAT33235.1 CH477669 EAT37499.1 DS231864 EDS40171.1 AXCN02001328 APCN01001091 AJWK01006101 JR037788 AEY57969.1 JXJN01025262 KQ981727 KYN36790.1 GL764129 EFZ18343.1 KQ761467 OAD57519.1 KQ435012 KZC13772.1 GEBQ01005030 JAT34947.1 KZ288347 PBC27583.1 KQ459604 KPI92082.1 KQ977791 KYM99838.1 ADTU01022285 KQ976703 KYM77121.1 DS235804 EEB17280.1 GECU01025797 JAS81909.1 GBHO01040503 GBRD01010876 GDHC01021911 JAG03101.1 JAG54948.1 JAP96717.1 GECZ01005592 JAS64177.1 GBBI01001665 JAC17047.1 GBGD01003070 JAC85819.1 GDHC01002293 JAQ16336.1 GDKW01003288 JAI53307.1 NNAY01001357 OXU24253.1 KQ435789 KOX74437.1 KK107894 EZA47310.1
OWR45307.1 BABH01025250 BABH01025251 BABH01025252 GAKP01012043 JAC46909.1 CH479179 EDW24679.1 GAKP01012045 JAC46907.1 CM000070 EAL27775.2 GDHF01002640 JAI49674.1 OUUW01000007 SPP83039.1 SPP83040.1 GANO01002093 JAB57778.1 GBXI01001720 JAD12572.1 GDHF01029322 JAI22992.1 SPP83041.1 GBXI01011741 GBXI01010512 GBXI01002214 JAD02551.1 JAD03780.1 JAD12078.1 CM000160 EDW98184.1 BT044310 ACH92375.1 AE014297 AY071042 CH902617 EDV43957.1 CM000364 EDX14101.1 CH954182 EDV53969.1 JRES01001300 KNC23785.1 CH480815 EDW43375.1 CH940650 EDW68336.1 UFQS01000205 UFQT01000205 SSX01329.1 SSX21709.1 GDAI01000070 JAI17533.1 CH933806 EDW16136.1 CH916373 EDV94984.1 CH964251 EDW83263.1 GAMC01001818 JAC04738.1 KRF83783.1 GFDL01007549 JAV27496.1 GGFM01005521 MBW26272.1 GGFJ01009608 MBW58749.1 GGFJ01009591 MBW58732.1 GAMD01000173 JAB01418.1 AXCM01002197 GFDF01000982 JAV13102.1 GL446193 EFN88296.1 AAAB01008846 EGK96401.1 GL440135 EFN66223.1 GGFK01010809 MBW44130.1 GEHC01000111 JAV47534.1 EAA06392.5 CCAG010012076 EZ423844 ADD20120.1 GGFL01005101 MBW69279.1 KQ982668 KYQ52511.1 GGFL01005023 MBW69201.1 ODYU01011413 SOQ57142.1 CH478321 EAT33235.1 CH477669 EAT37499.1 DS231864 EDS40171.1 AXCN02001328 APCN01001091 AJWK01006101 JR037788 AEY57969.1 JXJN01025262 KQ981727 KYN36790.1 GL764129 EFZ18343.1 KQ761467 OAD57519.1 KQ435012 KZC13772.1 GEBQ01005030 JAT34947.1 KZ288347 PBC27583.1 KQ459604 KPI92082.1 KQ977791 KYM99838.1 ADTU01022285 KQ976703 KYM77121.1 DS235804 EEB17280.1 GECU01025797 JAS81909.1 GBHO01040503 GBRD01010876 GDHC01021911 JAG03101.1 JAG54948.1 JAP96717.1 GECZ01005592 JAS64177.1 GBBI01001665 JAC17047.1 GBGD01003070 JAC85819.1 GDHC01002293 JAQ16336.1 GDKW01003288 JAI53307.1 NNAY01001357 OXU24253.1 KQ435789 KOX74437.1 KK107894 EZA47310.1
Proteomes
UP000218220
UP000283053
UP000007151
UP000005204
UP000008744
UP000001819
+ More
UP000268350 UP000095300 UP000002282 UP000000803 UP000007801 UP000000304 UP000008711 UP000095301 UP000037069 UP000001292 UP000008792 UP000009192 UP000192221 UP000001070 UP000007798 UP000075883 UP000008237 UP000007062 UP000069272 UP000000311 UP000075900 UP000075903 UP000078200 UP000092444 UP000075809 UP000075885 UP000008820 UP000092443 UP000002320 UP000075881 UP000075886 UP000076408 UP000075840 UP000076407 UP000092461 UP000092460 UP000075920 UP000075902 UP000078541 UP000002358 UP000076502 UP000242457 UP000053268 UP000078542 UP000005205 UP000005203 UP000078540 UP000009046 UP000075884 UP000215335 UP000053105 UP000053097
UP000268350 UP000095300 UP000002282 UP000000803 UP000007801 UP000000304 UP000008711 UP000095301 UP000037069 UP000001292 UP000008792 UP000009192 UP000192221 UP000001070 UP000007798 UP000075883 UP000008237 UP000007062 UP000069272 UP000000311 UP000075900 UP000075903 UP000078200 UP000092444 UP000075809 UP000075885 UP000008820 UP000092443 UP000002320 UP000075881 UP000075886 UP000076408 UP000075840 UP000076407 UP000092461 UP000092460 UP000075920 UP000075902 UP000078541 UP000002358 UP000076502 UP000242457 UP000053268 UP000078542 UP000005205 UP000005203 UP000078540 UP000009046 UP000075884 UP000215335 UP000053105 UP000053097
Interpro
IPR000387
TYR_PHOSPHATASE_dom
+ More
IPR020422 TYR_PHOSPHATASE_DUAL_dom
IPR000340 Dual-sp_phosphatase_cat-dom
IPR029021 Prot-tyrosine_phosphatase-like
IPR042165 PTPMT1
IPR016130 Tyr_Pase_AS
IPR003595 Tyr_Pase_cat
IPR027417 P-loop_NTPase
IPR023179 GTP-bd_ortho_bundle_sf
IPR030378 G_CP_dom
IPR006073 GTP_binding_domain
IPR020422 TYR_PHOSPHATASE_DUAL_dom
IPR000340 Dual-sp_phosphatase_cat-dom
IPR029021 Prot-tyrosine_phosphatase-like
IPR042165 PTPMT1
IPR016130 Tyr_Pase_AS
IPR003595 Tyr_Pase_cat
IPR027417 P-loop_NTPase
IPR023179 GTP-bd_ortho_bundle_sf
IPR030378 G_CP_dom
IPR006073 GTP_binding_domain
Gene 3D
ProteinModelPortal
A0A2A4JSU5
A0A2A4JTL5
A0A3S2TSF3
A0A212EV24
H9J5J1
A0A034VWS7
+ More
B4G513 A0A034VXS0 Q299C8 A0A0K8WF45 A0A3B0KAC6 A0A3B0K4W4 U5EYC1 A0A0A1XNP6 A0A0K8U8J9 A0A3B0KDF1 A0A1I8P7H9 A0A0A1WZ67 B4PNX4 B5RJ36 Q86BN8 B3LY53 B4R244 B3P8E5 A0A1I8M2Q8 A0A0L0BUN9 B4HFY2 B4M0F7 A0A336LYB7 Q86BN8-2 A0A0K8TTQ9 B4K5B0 A0A1W4U878 B4JT94 B4NG46 W8BZG4 A0A0Q9WP77 A0A1Q3FJ38 A0A2M3ZCP1 A0A2M4C097 A0A2M4C0B0 T1EB78 A0A182M0E1 A0A1L8E3E9 A0A1W4UL73 E2B7L8 F5HJE0 A0A182FY75 E2AK13 A0A2M4ATL7 A0A1W7R8S0 A0A182RPF8 A0A182UV99 A0A1A9VM33 Q7PRY8 D3TQZ2 A0A2M4CVI2 A0A151WXU2 A0A182PT42 A0A2M4CVH1 A0A2H1WVW9 Q16G69 Q16SP8 A0A1A9Y7H3 B0W9E6 A0A182K2H5 A0A182QGP3 A0A182Y7P3 A0A182IA25 A0A182WW65 A0A1B0CC35 V9ICB3 A0A1B0C7X0 A0A182W4F4 A0A182TTT3 A0A195F850 K7IR65 A0A1S4FQZ6 E9IM00 A0A310SGT1 A0A154PPI4 A0A1B6MG75 A0A2A3E741 A0A194PFM1 A0A195CIE6 A0A158NP97 A0A088A7J8 A0A195AY43 E0VV74 A0A1B6I4T3 A0A0A9W6U2 A0A182MZN7 A0A1B6GP30 A0A023F738 A0A069DWV9 A0A146MC61 A0A0P4VU03 A0A232F132 A0A0M9A041 A0A026VUG6
B4G513 A0A034VXS0 Q299C8 A0A0K8WF45 A0A3B0KAC6 A0A3B0K4W4 U5EYC1 A0A0A1XNP6 A0A0K8U8J9 A0A3B0KDF1 A0A1I8P7H9 A0A0A1WZ67 B4PNX4 B5RJ36 Q86BN8 B3LY53 B4R244 B3P8E5 A0A1I8M2Q8 A0A0L0BUN9 B4HFY2 B4M0F7 A0A336LYB7 Q86BN8-2 A0A0K8TTQ9 B4K5B0 A0A1W4U878 B4JT94 B4NG46 W8BZG4 A0A0Q9WP77 A0A1Q3FJ38 A0A2M3ZCP1 A0A2M4C097 A0A2M4C0B0 T1EB78 A0A182M0E1 A0A1L8E3E9 A0A1W4UL73 E2B7L8 F5HJE0 A0A182FY75 E2AK13 A0A2M4ATL7 A0A1W7R8S0 A0A182RPF8 A0A182UV99 A0A1A9VM33 Q7PRY8 D3TQZ2 A0A2M4CVI2 A0A151WXU2 A0A182PT42 A0A2M4CVH1 A0A2H1WVW9 Q16G69 Q16SP8 A0A1A9Y7H3 B0W9E6 A0A182K2H5 A0A182QGP3 A0A182Y7P3 A0A182IA25 A0A182WW65 A0A1B0CC35 V9ICB3 A0A1B0C7X0 A0A182W4F4 A0A182TTT3 A0A195F850 K7IR65 A0A1S4FQZ6 E9IM00 A0A310SGT1 A0A154PPI4 A0A1B6MG75 A0A2A3E741 A0A194PFM1 A0A195CIE6 A0A158NP97 A0A088A7J8 A0A195AY43 E0VV74 A0A1B6I4T3 A0A0A9W6U2 A0A182MZN7 A0A1B6GP30 A0A023F738 A0A069DWV9 A0A146MC61 A0A0P4VU03 A0A232F132 A0A0M9A041 A0A026VUG6
PDB
3RGO
E-value=2.18509e-32,
Score=343
Ontologies
GO
PANTHER
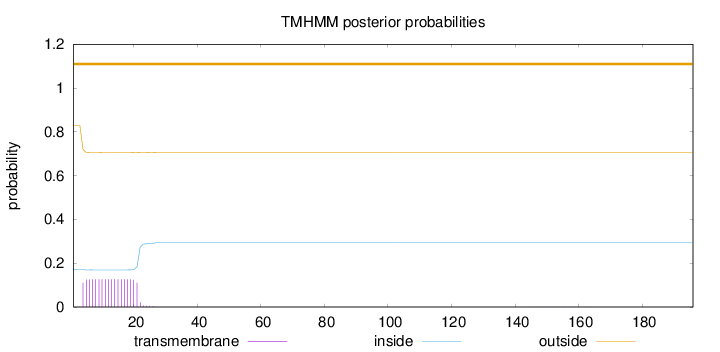
Topology
Subcellular location
Mitochondrion inner membrane
Length:
196
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
2.27948
Exp number, first 60 AAs:
2.27419
Total prob of N-in:
0.17070
outside
1 - 196
Population Genetic Test Statistics
Pi
194.634634
Theta
169.996375
Tajima's D
-1.564748
CLR
2.784894
CSRT
0.0514974251287436
Interpretation
Uncertain
