Gene
KWMTBOMO14977
Annotation
PREDICTED:_homeobox_protein_homothorax_[Bombyx_mori]
Full name
Homeobox protein homothorax
Alternative Name
Homeobox protein dorsotonals
Location in the cell
Mitochondrial Reliability : 1.076 Nuclear Reliability : 1.734
Sequence
CDS
ATGCCCGTGTTGCCAGGGTTCATAAACGCGAGACGTAGGATAGTCCAACCAATGATCGACCAATCAAATAGAGCAGTGTTCTACCCGTCAGTGTTCCCGCACGCCGGCCCCAGCGGCGCGTACAGTCCCGAGGCGACCATGGGCTACATGATGGACGGGCAGCAGATGATGCACAGGCCGCCCACAGACCCCGCCTTCCACCAGGGCTACGCGCACTACCCGGCCGAGTATTACGGACACCACCTCTAA
Protein
MPVLPGFINARRRIVQPMIDQSNRAVFYPSVFPHAGPSGAYSPEATMGYMMDGQQMMHRPPTDPAFHQGYAHYPAEYYGHHL
Summary
Description
All isoforms are required for patterning of the embryonic cuticle. Acts with exd to delimit the eye field and prevent inappropriate eye development. Isoforms that carry the homeodomain are required for proper localization of chordotonal organs within the peripheral nervous system and antennal identity; required to activate antennal-specific genes, such as sal and to repress the leg-like expression of dac. Necessary for the nuclear localization of the essential HOX cofactor, extradenticle (exd). Both necessary and sufficient for inner photoreceptors to adopt the polarization-sensitive 'dorsal rim area' (DRA) of the eye fate instead of the color-sensitive default state. This occurs by increasing rhabdomere size and uncoupling R7-R8 communication to allow both cells to express the same opsin rather than different ones as required for color vision.
Subunit
Interacts with exd; required for nuclear translocation of exd.
Similarity
Belongs to the TALE/MEIS homeobox family.
Keywords
Alternative splicing
Complete proteome
Developmental protein
DNA-binding
Homeobox
Nucleus
Reference proteome
Feature
chain Homeobox protein homothorax
splice variant In isoform H.
splice variant In isoform H.
Uniprot
A0A2H1WZX5
A0A2W1BJ35
A0A194PLV9
W6JHY5
A0A146M5Y1
A0A1Y1NAU0
+ More
A0A1Y1NBD1 Q70WC9 A0A1Y1NAU2 A0A1Y1NBY5 A0A0T6BAJ6 A0A3S2LTK6 A0A194RGQ9 A0A1U9WT16 A0A2R7WY80 A0A0C9RA77 A0A1B6CLX0 K7J253 A0A2S2QAQ8 T1H986 A0A088AB67 D8WUS7 A0A1B6H219 A0A2S2NNU6 A0A2S2R2C6 A0A1B6M0P5 A0A1B6MGF9 A0A2S2NQA2 J9K747 A0A2H8TJ49 A0A2H8TNK5 A0A1B6JR31 A0A1B6HQ36 A0A0C9RBF5 A0A1B6H5I3 A0A2M4BS80 A0A1B6GVZ1 A0A1B6IB01 A0A1B6GZW6 A0A1B6I2X4 A0A3L8D3I0 A0A2M3Z7T1 A0A0A9Z869 A0A2M4CSM0 T1GP55 A0A1Q3EUL3 A0A1W4U7A3 A0A1A9YE24 A0A0Q9WUH6 B4G681 I5ANA8 A0A023F5M7 A0A069DUQ5 A0A1A9ZWP5 B4JIJ5 A0A1A9WLB9 A0A3B0K029 A0A0Q9WPM1 B4K5K2 I5ANA3 A0A0R1E2S0 A0A0N8P183 B4QV74 B4PKQ5 B4HIS7 B3P1M7 B3LVY3 O46339 B4LWF2 A0A0Q9WYN2 I5ANA2 O46339-8 A0A182TYU8 A0A182VBJ9 A0A182XWI4 A0A182QPG7 O46339-2 A0A182M9S4 A0A182XR57 A0A1I8NMA7 A0A182WNJ1 A0A034WMA3 A0A0A1WYB3 B0WZR3 A0A1I8NMA8 A0A1I8N1L0 A0A034WQT8 A0A0A1XGW8 A0A182LQQ6 A0A1I8N1K7 A0A1I8N1J4 A0A084VRF8 A0A0L0C0P1 A0A0A9ZB98 A0A182IT36 Q7QK70 A0A0K8VYR1 A0A2M4CTY6 A0A0K8V4R9 W8BUB9
A0A1Y1NBD1 Q70WC9 A0A1Y1NAU2 A0A1Y1NBY5 A0A0T6BAJ6 A0A3S2LTK6 A0A194RGQ9 A0A1U9WT16 A0A2R7WY80 A0A0C9RA77 A0A1B6CLX0 K7J253 A0A2S2QAQ8 T1H986 A0A088AB67 D8WUS7 A0A1B6H219 A0A2S2NNU6 A0A2S2R2C6 A0A1B6M0P5 A0A1B6MGF9 A0A2S2NQA2 J9K747 A0A2H8TJ49 A0A2H8TNK5 A0A1B6JR31 A0A1B6HQ36 A0A0C9RBF5 A0A1B6H5I3 A0A2M4BS80 A0A1B6GVZ1 A0A1B6IB01 A0A1B6GZW6 A0A1B6I2X4 A0A3L8D3I0 A0A2M3Z7T1 A0A0A9Z869 A0A2M4CSM0 T1GP55 A0A1Q3EUL3 A0A1W4U7A3 A0A1A9YE24 A0A0Q9WUH6 B4G681 I5ANA8 A0A023F5M7 A0A069DUQ5 A0A1A9ZWP5 B4JIJ5 A0A1A9WLB9 A0A3B0K029 A0A0Q9WPM1 B4K5K2 I5ANA3 A0A0R1E2S0 A0A0N8P183 B4QV74 B4PKQ5 B4HIS7 B3P1M7 B3LVY3 O46339 B4LWF2 A0A0Q9WYN2 I5ANA2 O46339-8 A0A182TYU8 A0A182VBJ9 A0A182XWI4 A0A182QPG7 O46339-2 A0A182M9S4 A0A182XR57 A0A1I8NMA7 A0A182WNJ1 A0A034WMA3 A0A0A1WYB3 B0WZR3 A0A1I8NMA8 A0A1I8N1L0 A0A034WQT8 A0A0A1XGW8 A0A182LQQ6 A0A1I8N1K7 A0A1I8N1J4 A0A084VRF8 A0A0L0C0P1 A0A0A9ZB98 A0A182IT36 Q7QK70 A0A0K8VYR1 A0A2M4CTY6 A0A0K8V4R9 W8BUB9
Pubmed
EMBL
ODYU01012287
SOQ58516.1
KZ150023
PZC74868.1
KQ459604
KPI92090.1
+ More
AB841083 BAO48186.1 GDHC01004649 JAQ13980.1 GEZM01007633 JAV95032.1 GEZM01007636 JAV95029.1 AJ518941 CAD57735.1 GEZM01007634 JAV95031.1 GEZM01007637 JAV95028.1 LJIG01002582 KRT84358.1 RSAL01000216 RVE44127.1 KQ460205 KPJ17003.1 KX364396 AQY56477.1 KK855959 PTY24496.1 GBYB01009952 JAG79719.1 GEDC01022824 JAS14474.1 GGMS01005636 MBY74839.1 ACPB03016680 ACPB03016681 ACPB03016682 ACPB03016683 ACPB03016684 ACPB03016685 ACPB03016686 ACPB03016687 ACPB03016688 ACPB03016689 GU324093 ADI56665.1 GECZ01001046 JAS68723.1 GGMR01006188 MBY18807.1 GGMS01014891 MBY84094.1 GEBQ01010527 JAT29450.1 GEBQ01004992 JAT34985.1 GGMR01006679 MBY19298.1 ABLF02036466 GFXV01002331 MBW14136.1 GFXV01003948 MBW15753.1 GECU01033470 GECU01006101 JAS74236.1 JAT01606.1 GECU01030914 JAS76792.1 GBYB01004206 JAG73973.1 GECU01037741 JAS69965.1 GGFJ01006507 MBW55648.1 GECZ01013947 GECZ01003170 JAS55822.1 JAS66599.1 GECU01024555 GECU01023585 GECU01005424 GECU01003745 JAS83151.1 JAS84121.1 JAT02283.1 JAT03962.1 GECZ01003932 GECZ01001828 JAS65837.1 JAS67941.1 GECU01026433 GECU01009424 JAS81273.1 JAS98282.1 QOIP01000014 RLU15057.1 GGFM01003810 MBW24561.1 GBHO01003005 GBRD01001265 GDHC01003815 JAG40599.1 JAG64556.1 JAQ14814.1 GGFL01004135 MBW68313.1 CAQQ02055217 CAQQ02055218 CAQQ02055219 CAQQ02055220 CAQQ02055221 GFDL01016035 JAV19010.1 CH964251 KRF99692.1 CH479179 EDW23848.1 CM000070 EIM52443.2 GBBI01002245 JAC16467.1 GBGD01001482 JAC87407.1 CH916369 EDV93076.1 OUUW01000013 SPP87677.1 CH940650 KRF82820.1 CH933806 EDW14039.2 EIM52438.1 CM000160 KRK03375.1 CH902617 KPU79204.1 CM000364 EDX13480.1 EDW96814.1 CH480815 EDW42724.1 CH954181 EDV49626.1 EDV41516.1 KPU79200.1 KPU79202.1 AF026788 AF032865 AF035825 AF036584 AE014297 BT010238 BT024210 BT029117 AAB88514.1 AAB97169.1 AAC47759.1 AAN13474.1 ABC86272.1 EDW66589.2 KRG00821.1 EIM52437.2 AXCN02001551 AXCM01004490 GAKP01002276 JAC56676.1 GBXI01010904 JAD03388.1 DS232214 EDS37738.1 GAKP01002275 JAC56677.1 GBXI01004554 JAD09738.1 ATLV01015636 ATLV01015637 ATLV01015638 ATLV01015639 ATLV01015640 ATLV01015641 ATLV01015642 ATLV01015643 ATLV01015644 ATLV01015645 KE525025 KFB40552.1 JRES01001160 KNC25019.1 GBHO01003006 GBRD01001264 GDHC01017526 JAG40598.1 JAG64557.1 JAQ01103.1 AAAB01008799 EAA03775.5 GDHF01028057 GDHF01026463 GDHF01008307 JAI24257.1 JAI25851.1 JAI44007.1 GGFL01004634 MBW68812.1 GDHF01030431 GDHF01028885 GDHF01018443 JAI21883.1 JAI23429.1 JAI33871.1 GAMC01009649 JAB96906.1
AB841083 BAO48186.1 GDHC01004649 JAQ13980.1 GEZM01007633 JAV95032.1 GEZM01007636 JAV95029.1 AJ518941 CAD57735.1 GEZM01007634 JAV95031.1 GEZM01007637 JAV95028.1 LJIG01002582 KRT84358.1 RSAL01000216 RVE44127.1 KQ460205 KPJ17003.1 KX364396 AQY56477.1 KK855959 PTY24496.1 GBYB01009952 JAG79719.1 GEDC01022824 JAS14474.1 GGMS01005636 MBY74839.1 ACPB03016680 ACPB03016681 ACPB03016682 ACPB03016683 ACPB03016684 ACPB03016685 ACPB03016686 ACPB03016687 ACPB03016688 ACPB03016689 GU324093 ADI56665.1 GECZ01001046 JAS68723.1 GGMR01006188 MBY18807.1 GGMS01014891 MBY84094.1 GEBQ01010527 JAT29450.1 GEBQ01004992 JAT34985.1 GGMR01006679 MBY19298.1 ABLF02036466 GFXV01002331 MBW14136.1 GFXV01003948 MBW15753.1 GECU01033470 GECU01006101 JAS74236.1 JAT01606.1 GECU01030914 JAS76792.1 GBYB01004206 JAG73973.1 GECU01037741 JAS69965.1 GGFJ01006507 MBW55648.1 GECZ01013947 GECZ01003170 JAS55822.1 JAS66599.1 GECU01024555 GECU01023585 GECU01005424 GECU01003745 JAS83151.1 JAS84121.1 JAT02283.1 JAT03962.1 GECZ01003932 GECZ01001828 JAS65837.1 JAS67941.1 GECU01026433 GECU01009424 JAS81273.1 JAS98282.1 QOIP01000014 RLU15057.1 GGFM01003810 MBW24561.1 GBHO01003005 GBRD01001265 GDHC01003815 JAG40599.1 JAG64556.1 JAQ14814.1 GGFL01004135 MBW68313.1 CAQQ02055217 CAQQ02055218 CAQQ02055219 CAQQ02055220 CAQQ02055221 GFDL01016035 JAV19010.1 CH964251 KRF99692.1 CH479179 EDW23848.1 CM000070 EIM52443.2 GBBI01002245 JAC16467.1 GBGD01001482 JAC87407.1 CH916369 EDV93076.1 OUUW01000013 SPP87677.1 CH940650 KRF82820.1 CH933806 EDW14039.2 EIM52438.1 CM000160 KRK03375.1 CH902617 KPU79204.1 CM000364 EDX13480.1 EDW96814.1 CH480815 EDW42724.1 CH954181 EDV49626.1 EDV41516.1 KPU79200.1 KPU79202.1 AF026788 AF032865 AF035825 AF036584 AE014297 BT010238 BT024210 BT029117 AAB88514.1 AAB97169.1 AAC47759.1 AAN13474.1 ABC86272.1 EDW66589.2 KRG00821.1 EIM52437.2 AXCN02001551 AXCM01004490 GAKP01002276 JAC56676.1 GBXI01010904 JAD03388.1 DS232214 EDS37738.1 GAKP01002275 JAC56677.1 GBXI01004554 JAD09738.1 ATLV01015636 ATLV01015637 ATLV01015638 ATLV01015639 ATLV01015640 ATLV01015641 ATLV01015642 ATLV01015643 ATLV01015644 ATLV01015645 KE525025 KFB40552.1 JRES01001160 KNC25019.1 GBHO01003006 GBRD01001264 GDHC01017526 JAG40598.1 JAG64557.1 JAQ01103.1 AAAB01008799 EAA03775.5 GDHF01028057 GDHF01026463 GDHF01008307 JAI24257.1 JAI25851.1 JAI44007.1 GGFL01004634 MBW68812.1 GDHF01030431 GDHF01028885 GDHF01018443 JAI21883.1 JAI23429.1 JAI33871.1 GAMC01009649 JAB96906.1
Proteomes
UP000053268
UP000283053
UP000053240
UP000002358
UP000015103
UP000005203
+ More
UP000007819 UP000279307 UP000015102 UP000192221 UP000092443 UP000007798 UP000008744 UP000001819 UP000092445 UP000001070 UP000091820 UP000268350 UP000008792 UP000009192 UP000002282 UP000007801 UP000000304 UP000001292 UP000008711 UP000000803 UP000075902 UP000075903 UP000076408 UP000075886 UP000075883 UP000076407 UP000095300 UP000075920 UP000002320 UP000095301 UP000075882 UP000030765 UP000037069 UP000075880 UP000007062
UP000007819 UP000279307 UP000015102 UP000192221 UP000092443 UP000007798 UP000008744 UP000001819 UP000092445 UP000001070 UP000091820 UP000268350 UP000008792 UP000009192 UP000002282 UP000007801 UP000000304 UP000001292 UP000008711 UP000000803 UP000075902 UP000075903 UP000076408 UP000075886 UP000075883 UP000076407 UP000095300 UP000075920 UP000002320 UP000095301 UP000075882 UP000030765 UP000037069 UP000075880 UP000007062
Interpro
SUPFAM
SSF46689
SSF46689
ProteinModelPortal
A0A2H1WZX5
A0A2W1BJ35
A0A194PLV9
W6JHY5
A0A146M5Y1
A0A1Y1NAU0
+ More
A0A1Y1NBD1 Q70WC9 A0A1Y1NAU2 A0A1Y1NBY5 A0A0T6BAJ6 A0A3S2LTK6 A0A194RGQ9 A0A1U9WT16 A0A2R7WY80 A0A0C9RA77 A0A1B6CLX0 K7J253 A0A2S2QAQ8 T1H986 A0A088AB67 D8WUS7 A0A1B6H219 A0A2S2NNU6 A0A2S2R2C6 A0A1B6M0P5 A0A1B6MGF9 A0A2S2NQA2 J9K747 A0A2H8TJ49 A0A2H8TNK5 A0A1B6JR31 A0A1B6HQ36 A0A0C9RBF5 A0A1B6H5I3 A0A2M4BS80 A0A1B6GVZ1 A0A1B6IB01 A0A1B6GZW6 A0A1B6I2X4 A0A3L8D3I0 A0A2M3Z7T1 A0A0A9Z869 A0A2M4CSM0 T1GP55 A0A1Q3EUL3 A0A1W4U7A3 A0A1A9YE24 A0A0Q9WUH6 B4G681 I5ANA8 A0A023F5M7 A0A069DUQ5 A0A1A9ZWP5 B4JIJ5 A0A1A9WLB9 A0A3B0K029 A0A0Q9WPM1 B4K5K2 I5ANA3 A0A0R1E2S0 A0A0N8P183 B4QV74 B4PKQ5 B4HIS7 B3P1M7 B3LVY3 O46339 B4LWF2 A0A0Q9WYN2 I5ANA2 O46339-8 A0A182TYU8 A0A182VBJ9 A0A182XWI4 A0A182QPG7 O46339-2 A0A182M9S4 A0A182XR57 A0A1I8NMA7 A0A182WNJ1 A0A034WMA3 A0A0A1WYB3 B0WZR3 A0A1I8NMA8 A0A1I8N1L0 A0A034WQT8 A0A0A1XGW8 A0A182LQQ6 A0A1I8N1K7 A0A1I8N1J4 A0A084VRF8 A0A0L0C0P1 A0A0A9ZB98 A0A182IT36 Q7QK70 A0A0K8VYR1 A0A2M4CTY6 A0A0K8V4R9 W8BUB9
A0A1Y1NBD1 Q70WC9 A0A1Y1NAU2 A0A1Y1NBY5 A0A0T6BAJ6 A0A3S2LTK6 A0A194RGQ9 A0A1U9WT16 A0A2R7WY80 A0A0C9RA77 A0A1B6CLX0 K7J253 A0A2S2QAQ8 T1H986 A0A088AB67 D8WUS7 A0A1B6H219 A0A2S2NNU6 A0A2S2R2C6 A0A1B6M0P5 A0A1B6MGF9 A0A2S2NQA2 J9K747 A0A2H8TJ49 A0A2H8TNK5 A0A1B6JR31 A0A1B6HQ36 A0A0C9RBF5 A0A1B6H5I3 A0A2M4BS80 A0A1B6GVZ1 A0A1B6IB01 A0A1B6GZW6 A0A1B6I2X4 A0A3L8D3I0 A0A2M3Z7T1 A0A0A9Z869 A0A2M4CSM0 T1GP55 A0A1Q3EUL3 A0A1W4U7A3 A0A1A9YE24 A0A0Q9WUH6 B4G681 I5ANA8 A0A023F5M7 A0A069DUQ5 A0A1A9ZWP5 B4JIJ5 A0A1A9WLB9 A0A3B0K029 A0A0Q9WPM1 B4K5K2 I5ANA3 A0A0R1E2S0 A0A0N8P183 B4QV74 B4PKQ5 B4HIS7 B3P1M7 B3LVY3 O46339 B4LWF2 A0A0Q9WYN2 I5ANA2 O46339-8 A0A182TYU8 A0A182VBJ9 A0A182XWI4 A0A182QPG7 O46339-2 A0A182M9S4 A0A182XR57 A0A1I8NMA7 A0A182WNJ1 A0A034WMA3 A0A0A1WYB3 B0WZR3 A0A1I8NMA8 A0A1I8N1L0 A0A034WQT8 A0A0A1XGW8 A0A182LQQ6 A0A1I8N1K7 A0A1I8N1J4 A0A084VRF8 A0A0L0C0P1 A0A0A9ZB98 A0A182IT36 Q7QK70 A0A0K8VYR1 A0A2M4CTY6 A0A0K8V4R9 W8BUB9
PDB
3K2A
E-value=0.000352596,
Score=97
Ontologies
GO
GO:0003677
GO:0006355
GO:0005634
GO:0007383
GO:0090575
GO:0046982
GO:0045944
GO:0060323
GO:0007476
GO:0048735
GO:0042659
GO:0001158
GO:0000981
GO:0072002
GO:0032993
GO:0007480
GO:0001752
GO:0001742
GO:0007422
GO:2000497
GO:0007420
GO:0045664
GO:0007525
GO:0005737
GO:0001223
GO:0034504
GO:0090098
GO:0003700
GO:0035282
GO:0005667
GO:0007479
GO:0008134
GO:0006357
GO:0010092
GO:0009954
GO:0007380
GO:0005829
GO:0048749
GO:0005654
GO:0007432
GO:0035326
GO:0001654
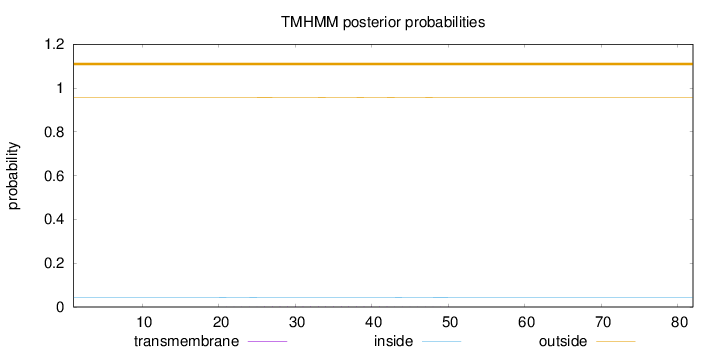
Topology
Subcellular location
Nucleus
Length:
82
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.03599
Exp number, first 60 AAs:
0.03599
Total prob of N-in:
0.04303
outside
1 - 82
Population Genetic Test Statistics
Pi
216.573717
Theta
168.904629
Tajima's D
0.369518
CLR
1.565168
CSRT
0.47647617619119
Interpretation
Uncertain
