Gene
KWMTBOMO14975
Pre Gene Modal
BGIBMGA000823
Annotation
putative_tumor_suppressor_protein_[Danaus_plexippus]
Location in the cell
Cytoplasmic Reliability : 1.456
Sequence
CDS
ATGCTCACGTCTGGAAGTCTACTTGGCAAAGGCTGCCAGGCGGCGACGAAGCTGGCCCATCTCGGGGAAGACGCCGAAAGGAATGCTTACATTTTGGGCGGCCATTTGGCTATAATGTGGCAACTATATTTAGACGTTAAAGACTTTTTCACCCACCCGTATTCATACTCTCTGGTTGGTGCTCCAGTAATCTTTGCCATTTGGGAATACCCAACTATTTACGGTCACATCTTGGAGTCGAAGTTGGAGAGGAAGCCCGTGGAATACAAGCAGTTGTATTACGCTGTAAGAAGTACAAGGTCTTTGGAGTATCTGTGCGTGTTTTTTGAGGAAGAACTGGCTTCAGTTTTGAAGTGTAGCGAGCGGTTCCCGGTGGATGACGCTAGGGTCGCGCTGCAGAAGATGGCGGTGACGATAAGAGATGAGGCCATGGGTTATATTGAAAAGTGA
Protein
MLTSGSLLGKGCQAATKLAHLGEDAERNAYILGGHLAIMWQLYLDVKDFFTHPYSYSLVGAPVIFAIWEYPTIYGHILESKLERKPVEYKQLYYAVRSTRSLEYLCVFFEEELASVLKCSERFPVDDARVALQKMAVTIRDEAMGYIEK
Summary
Similarity
Belongs to the FPP/GGPP synthase family.
Uniprot
H9IU93
A0A212F6F0
A0A0L7KNV2
A0A2A4JN95
A0A194RI51
A0A194PFM9
+ More
A0A2H1WZN3 A0A2W1BQ68 W8CD48 A0A0L0CNU2 A0A1A9X0N0 A0A1A9UJ74 A0A034WM77 B4N3V4 A0A1B0A6K5 A0A0K8VCT6 B4GUL6 A0A1I8PKI1 Q2M160 A0A1B0FBM7 A0A2A4J107 A0A1A9XJI0 A0A1B0AY55 B4J083 A0A212FDL4 A0A3B0JW05 B4PE82 B3NIU9 A0A0M4EDE5 B4IUK0 A0A0J9UQK4 Q9VP87 B4QJL9 B4IAI9 A0A1I8NIU7 T1PC98 B4LHG4 B3M6U9 A0A1B0CLL9 H9JT20 A0A1W4V9U8 Q17KC3 B4KWM6 A0A1I8QB34 A0A194RB96 U4U1Z8 U4TPP9 A0A0A1XPK4 A0A0L7LLB7 A0A194Q7W4 A0A2M4C5H7 A0A336K210 N6SVZ1 A0A1J1I8A6 A0A2M4BPK6 A0A2M4BP09 A0A084W0S5 A0A084W0S6 A0A2M4BP46 A0A1W4WH88 A0A182TZT9 A0A182GYG7 B0XA19 A0A2M4BN91 A0A2M4BN23 A0A1Y1N0V5 V5UUA7 B0XA18 A0A194PFE8 A0A182JBB9 A0A182TB49 A0A182P5L9 A0A1Y9HEW6 A0A2S2QGI7 A0A182GYG6 J9K7E9 A0A2H8TCK0 M9VRD0 A0A2M4AET4 A0A182V0X2 A0A2M4AER4 A0A194RHT9 A0A182YCX5 A0A2S2P200 Q7Q671 W5JCC3 A0A182KPF9 A0A182I1R4 A0A2S2QNZ5 A0A182NIH7 Q17KC4 A0A182FCQ0 A0A1S4EZP3 A0A084VAU1 A0A336LS10 A0A182S7M9 A0A1Q3FCN9 A0A182GBA4 A0A182K9F5 A0A182ML90
A0A2H1WZN3 A0A2W1BQ68 W8CD48 A0A0L0CNU2 A0A1A9X0N0 A0A1A9UJ74 A0A034WM77 B4N3V4 A0A1B0A6K5 A0A0K8VCT6 B4GUL6 A0A1I8PKI1 Q2M160 A0A1B0FBM7 A0A2A4J107 A0A1A9XJI0 A0A1B0AY55 B4J083 A0A212FDL4 A0A3B0JW05 B4PE82 B3NIU9 A0A0M4EDE5 B4IUK0 A0A0J9UQK4 Q9VP87 B4QJL9 B4IAI9 A0A1I8NIU7 T1PC98 B4LHG4 B3M6U9 A0A1B0CLL9 H9JT20 A0A1W4V9U8 Q17KC3 B4KWM6 A0A1I8QB34 A0A194RB96 U4U1Z8 U4TPP9 A0A0A1XPK4 A0A0L7LLB7 A0A194Q7W4 A0A2M4C5H7 A0A336K210 N6SVZ1 A0A1J1I8A6 A0A2M4BPK6 A0A2M4BP09 A0A084W0S5 A0A084W0S6 A0A2M4BP46 A0A1W4WH88 A0A182TZT9 A0A182GYG7 B0XA19 A0A2M4BN91 A0A2M4BN23 A0A1Y1N0V5 V5UUA7 B0XA18 A0A194PFE8 A0A182JBB9 A0A182TB49 A0A182P5L9 A0A1Y9HEW6 A0A2S2QGI7 A0A182GYG6 J9K7E9 A0A2H8TCK0 M9VRD0 A0A2M4AET4 A0A182V0X2 A0A2M4AER4 A0A194RHT9 A0A182YCX5 A0A2S2P200 Q7Q671 W5JCC3 A0A182KPF9 A0A182I1R4 A0A2S2QNZ5 A0A182NIH7 Q17KC4 A0A182FCQ0 A0A1S4EZP3 A0A084VAU1 A0A336LS10 A0A182S7M9 A0A1Q3FCN9 A0A182GBA4 A0A182K9F5 A0A182ML90
Pubmed
19121390
22118469
26227816
26354079
28756777
24495485
+ More
26108605 25348373 17994087 15632085 17550304 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25315136 17510324 23537049 25830018 24438588 26483478 28004739 24246678 25244985 12364791 14747013 17210077 20920257 23761445 20966253
26108605 25348373 17994087 15632085 17550304 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25315136 17510324 23537049 25830018 24438588 26483478 28004739 24246678 25244985 12364791 14747013 17210077 20920257 23761445 20966253
EMBL
BABH01034256
AGBW02010046
OWR49298.1
JTDY01008044
KOB64780.1
NWSH01000929
+ More
PCG73507.1 KQ460205 KPJ17005.1 KQ459604 KPI92092.1 ODYU01012287 SOQ58513.1 KZ150023 PZC74870.1 GAMC01002571 JAC03985.1 JRES01000128 KNC33931.1 GAKP01003492 GAKP01003490 JAC55462.1 CH964095 EDW79309.2 GDHF01015616 JAI36698.1 CH479191 EDW26299.1 CH379069 EAL30715.1 CCAG010008960 NWSH01004165 PCG65416.1 JXJN01005555 CH916366 EDV95684.1 AGBW02009028 OWR51824.1 OUUW01000002 SPP76901.1 CM000159 EDW95095.1 CH954178 EDV52595.1 CP012525 ALC43608.1 CH891933 EDX00064.1 CM002912 KMZ00916.1 AE014296 AY051597 AAF51668.1 AAK93021.1 CM000363 EDX11319.1 CH480826 EDW44302.1 KA646386 AFP61015.1 CH940647 EDW68494.1 CH902618 EDV39785.1 AJWK01017431 BABH01030708 CH477226 EAT47124.1 CH933809 EDW17473.1 KQ460416 KPJ14892.1 KB631048 KB631049 ERL84043.1 ERL84045.1 KB630114 ERL83404.1 GBXI01016343 GBXI01001023 JAC97948.1 JAD13269.1 JTDY01000645 KOB76353.1 KQ459324 KPJ01613.1 GGFJ01011441 MBW60582.1 UFQS01000067 UFQT01000067 SSW98956.1 SSX19338.1 APGK01054216 APGK01054217 APGK01054218 KB741248 KB631854 ENN71929.1 ERL86744.1 CVRI01000043 CRK95970.1 GGFJ01005859 MBW55000.1 GGFJ01005644 MBW54785.1 ATLV01019157 KE525263 KFB43819.1 KFB43820.1 GGFJ01005580 MBW54721.1 JXUM01021119 JXUM01021120 JXUM01021121 KQ560591 KXJ81758.1 DS232561 EDS43445.1 GGFJ01005270 MBW54411.1 GGFJ01005271 MBW54412.1 GEZM01016911 JAV91048.1 KF673862 AHB81558.1 EDS43444.1 KPI92091.1 GGMS01007608 MBY76811.1 KXJ81757.1 ABLF02016144 GFXV01000039 MBW11844.1 KC431244 AGJ83980.1 GGFK01005952 MBW39273.1 GGFK01005960 MBW39281.1 KPJ17004.1 GGMR01010785 MBY23404.1 AAAB01008960 EAA11709.4 ADMH02001837 ETN60973.1 GGMS01010286 MBY79489.1 EAT47123.1 ATLV01004460 ATLV01004461 KE524214 KFB35085.1 SSX19359.1 GFDL01009756 JAV25289.1 JXUM01241826 KQ635302 KXJ62490.1 AXCM01006428
PCG73507.1 KQ460205 KPJ17005.1 KQ459604 KPI92092.1 ODYU01012287 SOQ58513.1 KZ150023 PZC74870.1 GAMC01002571 JAC03985.1 JRES01000128 KNC33931.1 GAKP01003492 GAKP01003490 JAC55462.1 CH964095 EDW79309.2 GDHF01015616 JAI36698.1 CH479191 EDW26299.1 CH379069 EAL30715.1 CCAG010008960 NWSH01004165 PCG65416.1 JXJN01005555 CH916366 EDV95684.1 AGBW02009028 OWR51824.1 OUUW01000002 SPP76901.1 CM000159 EDW95095.1 CH954178 EDV52595.1 CP012525 ALC43608.1 CH891933 EDX00064.1 CM002912 KMZ00916.1 AE014296 AY051597 AAF51668.1 AAK93021.1 CM000363 EDX11319.1 CH480826 EDW44302.1 KA646386 AFP61015.1 CH940647 EDW68494.1 CH902618 EDV39785.1 AJWK01017431 BABH01030708 CH477226 EAT47124.1 CH933809 EDW17473.1 KQ460416 KPJ14892.1 KB631048 KB631049 ERL84043.1 ERL84045.1 KB630114 ERL83404.1 GBXI01016343 GBXI01001023 JAC97948.1 JAD13269.1 JTDY01000645 KOB76353.1 KQ459324 KPJ01613.1 GGFJ01011441 MBW60582.1 UFQS01000067 UFQT01000067 SSW98956.1 SSX19338.1 APGK01054216 APGK01054217 APGK01054218 KB741248 KB631854 ENN71929.1 ERL86744.1 CVRI01000043 CRK95970.1 GGFJ01005859 MBW55000.1 GGFJ01005644 MBW54785.1 ATLV01019157 KE525263 KFB43819.1 KFB43820.1 GGFJ01005580 MBW54721.1 JXUM01021119 JXUM01021120 JXUM01021121 KQ560591 KXJ81758.1 DS232561 EDS43445.1 GGFJ01005270 MBW54411.1 GGFJ01005271 MBW54412.1 GEZM01016911 JAV91048.1 KF673862 AHB81558.1 EDS43444.1 KPI92091.1 GGMS01007608 MBY76811.1 KXJ81757.1 ABLF02016144 GFXV01000039 MBW11844.1 KC431244 AGJ83980.1 GGFK01005952 MBW39273.1 GGFK01005960 MBW39281.1 KPJ17004.1 GGMR01010785 MBY23404.1 AAAB01008960 EAA11709.4 ADMH02001837 ETN60973.1 GGMS01010286 MBY79489.1 EAT47123.1 ATLV01004460 ATLV01004461 KE524214 KFB35085.1 SSX19359.1 GFDL01009756 JAV25289.1 JXUM01241826 KQ635302 KXJ62490.1 AXCM01006428
Proteomes
UP000005204
UP000007151
UP000037510
UP000218220
UP000053240
UP000053268
+ More
UP000037069 UP000091820 UP000078200 UP000007798 UP000092445 UP000008744 UP000095300 UP000001819 UP000092444 UP000092443 UP000092460 UP000001070 UP000268350 UP000002282 UP000008711 UP000092553 UP000000803 UP000000304 UP000001292 UP000095301 UP000008792 UP000007801 UP000092461 UP000192221 UP000008820 UP000009192 UP000030742 UP000019118 UP000183832 UP000030765 UP000192223 UP000075902 UP000069940 UP000249989 UP000002320 UP000075880 UP000075901 UP000075885 UP000075900 UP000007819 UP000075903 UP000076408 UP000007062 UP000000673 UP000075882 UP000075884 UP000069272 UP000075881 UP000075883
UP000037069 UP000091820 UP000078200 UP000007798 UP000092445 UP000008744 UP000095300 UP000001819 UP000092444 UP000092443 UP000092460 UP000001070 UP000268350 UP000002282 UP000008711 UP000092553 UP000000803 UP000000304 UP000001292 UP000095301 UP000008792 UP000007801 UP000092461 UP000192221 UP000008820 UP000009192 UP000030742 UP000019118 UP000183832 UP000030765 UP000192223 UP000075902 UP000069940 UP000249989 UP000002320 UP000075880 UP000075901 UP000075885 UP000075900 UP000007819 UP000075903 UP000076408 UP000007062 UP000000673 UP000075882 UP000075884 UP000069272 UP000075881 UP000075883
Pfam
PF00348 polyprenyl_synt
SUPFAM
SSF48576
SSF48576
Gene 3D
ProteinModelPortal
H9IU93
A0A212F6F0
A0A0L7KNV2
A0A2A4JN95
A0A194RI51
A0A194PFM9
+ More
A0A2H1WZN3 A0A2W1BQ68 W8CD48 A0A0L0CNU2 A0A1A9X0N0 A0A1A9UJ74 A0A034WM77 B4N3V4 A0A1B0A6K5 A0A0K8VCT6 B4GUL6 A0A1I8PKI1 Q2M160 A0A1B0FBM7 A0A2A4J107 A0A1A9XJI0 A0A1B0AY55 B4J083 A0A212FDL4 A0A3B0JW05 B4PE82 B3NIU9 A0A0M4EDE5 B4IUK0 A0A0J9UQK4 Q9VP87 B4QJL9 B4IAI9 A0A1I8NIU7 T1PC98 B4LHG4 B3M6U9 A0A1B0CLL9 H9JT20 A0A1W4V9U8 Q17KC3 B4KWM6 A0A1I8QB34 A0A194RB96 U4U1Z8 U4TPP9 A0A0A1XPK4 A0A0L7LLB7 A0A194Q7W4 A0A2M4C5H7 A0A336K210 N6SVZ1 A0A1J1I8A6 A0A2M4BPK6 A0A2M4BP09 A0A084W0S5 A0A084W0S6 A0A2M4BP46 A0A1W4WH88 A0A182TZT9 A0A182GYG7 B0XA19 A0A2M4BN91 A0A2M4BN23 A0A1Y1N0V5 V5UUA7 B0XA18 A0A194PFE8 A0A182JBB9 A0A182TB49 A0A182P5L9 A0A1Y9HEW6 A0A2S2QGI7 A0A182GYG6 J9K7E9 A0A2H8TCK0 M9VRD0 A0A2M4AET4 A0A182V0X2 A0A2M4AER4 A0A194RHT9 A0A182YCX5 A0A2S2P200 Q7Q671 W5JCC3 A0A182KPF9 A0A182I1R4 A0A2S2QNZ5 A0A182NIH7 Q17KC4 A0A182FCQ0 A0A1S4EZP3 A0A084VAU1 A0A336LS10 A0A182S7M9 A0A1Q3FCN9 A0A182GBA4 A0A182K9F5 A0A182ML90
A0A2H1WZN3 A0A2W1BQ68 W8CD48 A0A0L0CNU2 A0A1A9X0N0 A0A1A9UJ74 A0A034WM77 B4N3V4 A0A1B0A6K5 A0A0K8VCT6 B4GUL6 A0A1I8PKI1 Q2M160 A0A1B0FBM7 A0A2A4J107 A0A1A9XJI0 A0A1B0AY55 B4J083 A0A212FDL4 A0A3B0JW05 B4PE82 B3NIU9 A0A0M4EDE5 B4IUK0 A0A0J9UQK4 Q9VP87 B4QJL9 B4IAI9 A0A1I8NIU7 T1PC98 B4LHG4 B3M6U9 A0A1B0CLL9 H9JT20 A0A1W4V9U8 Q17KC3 B4KWM6 A0A1I8QB34 A0A194RB96 U4U1Z8 U4TPP9 A0A0A1XPK4 A0A0L7LLB7 A0A194Q7W4 A0A2M4C5H7 A0A336K210 N6SVZ1 A0A1J1I8A6 A0A2M4BPK6 A0A2M4BP09 A0A084W0S5 A0A084W0S6 A0A2M4BP46 A0A1W4WH88 A0A182TZT9 A0A182GYG7 B0XA19 A0A2M4BN91 A0A2M4BN23 A0A1Y1N0V5 V5UUA7 B0XA18 A0A194PFE8 A0A182JBB9 A0A182TB49 A0A182P5L9 A0A1Y9HEW6 A0A2S2QGI7 A0A182GYG6 J9K7E9 A0A2H8TCK0 M9VRD0 A0A2M4AET4 A0A182V0X2 A0A2M4AER4 A0A194RHT9 A0A182YCX5 A0A2S2P200 Q7Q671 W5JCC3 A0A182KPF9 A0A182I1R4 A0A2S2QNZ5 A0A182NIH7 Q17KC4 A0A182FCQ0 A0A1S4EZP3 A0A084VAU1 A0A336LS10 A0A182S7M9 A0A1Q3FCN9 A0A182GBA4 A0A182K9F5 A0A182ML90
Ontologies
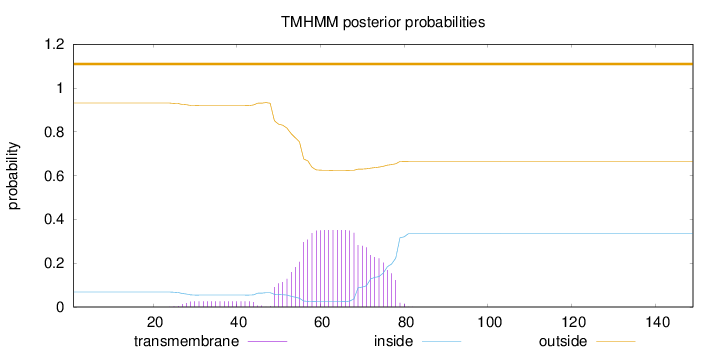
Topology
Length:
149
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
8.08968999999998
Exp number, first 60 AAs:
3.09134
Total prob of N-in:
0.06874
outside
1 - 149
Population Genetic Test Statistics
Pi
200.40117
Theta
151.494466
Tajima's D
0.957903
CLR
0
CSRT
0.650517474126294
Interpretation
Uncertain
