Gene
KWMTBOMO14970
Pre Gene Modal
BGIBMGA000821
Annotation
PREDICTED:_radial_spoke_head_protein_6_homolog_A_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.96
Sequence
CDS
ATGTTGTGTTACCTTAGCGAACAGTGGATCTGTCTCCCGGATGTAACTCCTGATCAGATCAGAGTGGCCCGCCTCACTGTGCGTTGTATGACCGGAAATCTAGATGCTGAGGTTCAATCGTTTCCGCCGTTTGAGGGAACAGAGCGGAATTATCTGCGCGCTCAACTAGCTCGGATCCAAGCCGGCACGTCCGTGTCTCCTCAGGGCTTCTACACTTTTGGTTCTGGTGAAGAAGAAGATATTGATTTGGAGGAAGGCGGGGGAGATTTAAATTTCAACCCCAATCCGTTTTACCACGGCCACTCGATGAAGGACCTGCTGGACTCGAGCCTCACGTACTGGGTGCATCATGGACGACATATACTGAAACAGGGTAGGACTCTCTGGTGGAATCCTAATGCTGGAATGGAGTTTCTGTACTCTAGAACGGCTATGAGTAAGAAAATAACCTATGCCAAGCCACTAGCGAGGCAGTTAGAGAAGGCAGCGTCAATAATTATGGAGGAAGAAGAAGCACAATGCCGGAAATAA
Protein
MLCYLSEQWICLPDVTPDQIRVARLTVRCMTGNLDAEVQSFPPFEGTERNYLRAQLARIQAGTSVSPQGFYTFGSGEEEDIDLEEGGGDLNFNPNPFYHGHSMKDLLDSSLTYWVHHGRHILKQGRTLWWNPNAGMEFLYSRTAMSKKITYAKPLARQLEKAASIIMEEEEAQCRK
Summary
Uniprot
A0A212F6E7
A0A2W1B7R5
A0A2A4JFB4
A0A194RHU4
A0A194PLW4
A0A2H1VJ98
+ More
A0A154P3P6 A0A0L7KW62 E0W087 D6WQE3 A0A158NCR3 A0A075AJR4 A0A1S8WLA3 A0A151I2R2 A0A310SUH3 A0A183B2V7 A0A3R7GRT6 A0A151XCQ9 E2A4D0 G7Y5T9 A0A151J996 A0A0L7QY67 H2KRA8 A0A1W4WQT0 F4WYR7 A0A3Q0KGB2 A0A1I8HZC7 A0A1I8GZ50 A0A1I8FY27 G4V5J5 A0A074YZD8 A0A088A5U1 A0A195CPE1 A0A183LKS7 A0A2A3EIV7 A0A183NYC3 A0A183K5E4 E4X3B0 T1F7C2 A0A3L8D4R2 A0A1I8INK1 H3BAM5 A0A026W6Y8 A0A095B2K7 A0A0B7B924 E9IM35 A0A0B7B639 A0A0N0BDF6 A0A3S1ASK2 O46178 W4YGB6 V4ABQ0 A0A0B7B9S3 A0A2G8JST0 R7UDR4 H2ZH03 A0A2Y9DDF4 A0A1S3D3V4 A0A2C9K7T3 A0A1B6K2V7 C4B8X4 A0A341CEV0 A0A093R2Y8 S7MG66 F7FU36 C3ZMT5 A0A091JCA6 A0A1W3JEG2 G1SY17 A0A183LJ54 A0A183K760 G1PTD3 U3KPB6 A0A095A255 A0A183RP67 A0A093J9S8 A0A2U3XT83 A0A3Q7SRB7 A0A2F0BCW9 A0A1S3IY04 G4VCB4 A0A087QR25 A0A091WAZ0 A0A2U3V606 A0A1S3I1H4 M3X9Z3 A0A3Q0KMB6 F6UPL6 A0A3M6UUI2 F6Q808 H0UU47 A0A340X040 A0A2Y9FD02 A0A287D6C0 A0A2U3ZDC7 G5AMZ9
A0A154P3P6 A0A0L7KW62 E0W087 D6WQE3 A0A158NCR3 A0A075AJR4 A0A1S8WLA3 A0A151I2R2 A0A310SUH3 A0A183B2V7 A0A3R7GRT6 A0A151XCQ9 E2A4D0 G7Y5T9 A0A151J996 A0A0L7QY67 H2KRA8 A0A1W4WQT0 F4WYR7 A0A3Q0KGB2 A0A1I8HZC7 A0A1I8GZ50 A0A1I8FY27 G4V5J5 A0A074YZD8 A0A088A5U1 A0A195CPE1 A0A183LKS7 A0A2A3EIV7 A0A183NYC3 A0A183K5E4 E4X3B0 T1F7C2 A0A3L8D4R2 A0A1I8INK1 H3BAM5 A0A026W6Y8 A0A095B2K7 A0A0B7B924 E9IM35 A0A0B7B639 A0A0N0BDF6 A0A3S1ASK2 O46178 W4YGB6 V4ABQ0 A0A0B7B9S3 A0A2G8JST0 R7UDR4 H2ZH03 A0A2Y9DDF4 A0A1S3D3V4 A0A2C9K7T3 A0A1B6K2V7 C4B8X4 A0A341CEV0 A0A093R2Y8 S7MG66 F7FU36 C3ZMT5 A0A091JCA6 A0A1W3JEG2 G1SY17 A0A183LJ54 A0A183K760 G1PTD3 U3KPB6 A0A095A255 A0A183RP67 A0A093J9S8 A0A2U3XT83 A0A3Q7SRB7 A0A2F0BCW9 A0A1S3IY04 G4VCB4 A0A087QR25 A0A091WAZ0 A0A2U3V606 A0A1S3I1H4 M3X9Z3 A0A3Q0KMB6 F6UPL6 A0A3M6UUI2 F6Q808 H0UU47 A0A340X040 A0A2Y9FD02 A0A287D6C0 A0A2U3ZDC7 G5AMZ9
Pubmed
22118469
28756777
26354079
26227816
20566863
18362917
+ More
19820115 21347285 20798317 22023798 21719571 22253936 26392545 21097902 23254933 30249741 9215903 24508170 22246508 21282665 9450971 29023486 15562597 12481130 15114417 19527718 18464734 18563158 21993624 17975172 16341006 30382153 19892987 21993625
19820115 21347285 20798317 22023798 21719571 22253936 26392545 21097902 23254933 30249741 9215903 24508170 22246508 21282665 9450971 29023486 15562597 12481130 15114417 19527718 18464734 18563158 21993624 17975172 16341006 30382153 19892987 21993625
EMBL
AGBW02010046
OWR49294.1
KZ150248
PZC71842.1
NWSH01001601
PCG70747.1
+ More
KQ460205 KPJ17009.1 KQ459604 KPI92095.1 ODYU01002874 SOQ40898.1 KQ434809 KZC06536.1 JTDY01005195 KOB67269.1 DS235857 EEB19043.1 KQ971354 EFA06085.1 ADTU01011560 ADTU01011561 KL596621 KER33839.1 KV906164 OON15227.1 KQ976508 KYM82746.1 KQ760329 OAD60867.1 UZAN01055346 VDP90814.1 NIRI01002532 RJW60925.1 KQ982298 KYQ58129.1 GL436635 EFN71677.1 DF142882 GAA48325.1 KQ979425 KYN21616.1 KQ414694 KOC63553.1 DF143135 GAA36169.2 GL888463 EGI60571.1 NIVC01000777 NIVC01000776 PAA76946.1 PAA76963.1 NIVC01000387 PAA84117.1 HE601624 CCD75639.1 KL597101 KER20038.1 KQ977565 KYN01974.1 UZAI01001401 VDO61528.1 KZ288229 PBC31705.1 UZAL01027998 VDP37586.1 UZAK01033649 VDP38917.1 FN653023 CBY18114.1 AMQM01004758 KB096676 ESO03028.1 QOIP01000014 RLU15131.1 AFYH01070735 AFYH01070736 KK107366 EZA51872.1 KL251872 KGB41421.1 HACG01041961 CEK88826.1 GL764129 EFZ18520.1 HACG01041954 CEK88819.1 KQ435877 KOX70098.1 RQTK01001303 RUS70929.1 U73123 AAC38955.1 AAGJ04104728 AAGJ04104729 KB201847 ESO94242.1 HACG01041955 CEK88820.1 MRZV01001315 PIK38778.1 AMQN01008104 KB302198 ELU04530.1 GECU01001921 JAT05786.1 EAAA01000632 AB501298 BAH59283.1 KL436421 KFW93010.1 KE161334 EPQ03174.1 GG666648 EEN46075.1 KK501736 KFP17433.1 AAGW02029260 AAGW02029261 AAGW02029262 UZAI01001152 VDO59249.1 AAPE02018719 KL251636 KGB40907.1 KL217263 KFV75669.1 NTJE010141188 MBW00854.1 HE601625 CCD77539.1 KL225822 KFM03679.1 KK735119 KFR12704.1 AANG04000169 AAEX03000382 RCHS01000676 RMX57332.1 AAKN02003415 AGTP01123348 AGTP01123349 AGTP01123350 JH166111 EHA98409.1
KQ460205 KPJ17009.1 KQ459604 KPI92095.1 ODYU01002874 SOQ40898.1 KQ434809 KZC06536.1 JTDY01005195 KOB67269.1 DS235857 EEB19043.1 KQ971354 EFA06085.1 ADTU01011560 ADTU01011561 KL596621 KER33839.1 KV906164 OON15227.1 KQ976508 KYM82746.1 KQ760329 OAD60867.1 UZAN01055346 VDP90814.1 NIRI01002532 RJW60925.1 KQ982298 KYQ58129.1 GL436635 EFN71677.1 DF142882 GAA48325.1 KQ979425 KYN21616.1 KQ414694 KOC63553.1 DF143135 GAA36169.2 GL888463 EGI60571.1 NIVC01000777 NIVC01000776 PAA76946.1 PAA76963.1 NIVC01000387 PAA84117.1 HE601624 CCD75639.1 KL597101 KER20038.1 KQ977565 KYN01974.1 UZAI01001401 VDO61528.1 KZ288229 PBC31705.1 UZAL01027998 VDP37586.1 UZAK01033649 VDP38917.1 FN653023 CBY18114.1 AMQM01004758 KB096676 ESO03028.1 QOIP01000014 RLU15131.1 AFYH01070735 AFYH01070736 KK107366 EZA51872.1 KL251872 KGB41421.1 HACG01041961 CEK88826.1 GL764129 EFZ18520.1 HACG01041954 CEK88819.1 KQ435877 KOX70098.1 RQTK01001303 RUS70929.1 U73123 AAC38955.1 AAGJ04104728 AAGJ04104729 KB201847 ESO94242.1 HACG01041955 CEK88820.1 MRZV01001315 PIK38778.1 AMQN01008104 KB302198 ELU04530.1 GECU01001921 JAT05786.1 EAAA01000632 AB501298 BAH59283.1 KL436421 KFW93010.1 KE161334 EPQ03174.1 GG666648 EEN46075.1 KK501736 KFP17433.1 AAGW02029260 AAGW02029261 AAGW02029262 UZAI01001152 VDO59249.1 AAPE02018719 KL251636 KGB40907.1 KL217263 KFV75669.1 NTJE010141188 MBW00854.1 HE601625 CCD77539.1 KL225822 KFM03679.1 KK735119 KFR12704.1 AANG04000169 AAEX03000382 RCHS01000676 RMX57332.1 AAKN02003415 AGTP01123348 AGTP01123349 AGTP01123350 JH166111 EHA98409.1
Proteomes
UP000007151
UP000218220
UP000053240
UP000053268
UP000076502
UP000037510
+ More
UP000009046 UP000007266 UP000005205 UP000078540 UP000050740 UP000075809 UP000000311 UP000078492 UP000053825 UP000192223 UP000007755 UP000008854 UP000095280 UP000215902 UP000005203 UP000078542 UP000050790 UP000277204 UP000242457 UP000050791 UP000050789 UP000015101 UP000279307 UP000008672 UP000053097 UP000053105 UP000271974 UP000007110 UP000030746 UP000230750 UP000014760 UP000007875 UP000248480 UP000079169 UP000076420 UP000008144 UP000252040 UP000002279 UP000001554 UP000053119 UP000001811 UP000001074 UP000050792 UP000053875 UP000245341 UP000286640 UP000085678 UP000053286 UP000053605 UP000245320 UP000011712 UP000002254 UP000275408 UP000002281 UP000005447 UP000265300 UP000248484 UP000005215 UP000245340 UP000006813
UP000009046 UP000007266 UP000005205 UP000078540 UP000050740 UP000075809 UP000000311 UP000078492 UP000053825 UP000192223 UP000007755 UP000008854 UP000095280 UP000215902 UP000005203 UP000078542 UP000050790 UP000277204 UP000242457 UP000050791 UP000050789 UP000015101 UP000279307 UP000008672 UP000053097 UP000053105 UP000271974 UP000007110 UP000030746 UP000230750 UP000014760 UP000007875 UP000248480 UP000079169 UP000076420 UP000008144 UP000252040 UP000002279 UP000001554 UP000053119 UP000001811 UP000001074 UP000050792 UP000053875 UP000245341 UP000286640 UP000085678 UP000053286 UP000053605 UP000245320 UP000011712 UP000002254 UP000275408 UP000002281 UP000005447 UP000265300 UP000248484 UP000005215 UP000245340 UP000006813
Interpro
Gene 3D
ProteinModelPortal
A0A212F6E7
A0A2W1B7R5
A0A2A4JFB4
A0A194RHU4
A0A194PLW4
A0A2H1VJ98
+ More
A0A154P3P6 A0A0L7KW62 E0W087 D6WQE3 A0A158NCR3 A0A075AJR4 A0A1S8WLA3 A0A151I2R2 A0A310SUH3 A0A183B2V7 A0A3R7GRT6 A0A151XCQ9 E2A4D0 G7Y5T9 A0A151J996 A0A0L7QY67 H2KRA8 A0A1W4WQT0 F4WYR7 A0A3Q0KGB2 A0A1I8HZC7 A0A1I8GZ50 A0A1I8FY27 G4V5J5 A0A074YZD8 A0A088A5U1 A0A195CPE1 A0A183LKS7 A0A2A3EIV7 A0A183NYC3 A0A183K5E4 E4X3B0 T1F7C2 A0A3L8D4R2 A0A1I8INK1 H3BAM5 A0A026W6Y8 A0A095B2K7 A0A0B7B924 E9IM35 A0A0B7B639 A0A0N0BDF6 A0A3S1ASK2 O46178 W4YGB6 V4ABQ0 A0A0B7B9S3 A0A2G8JST0 R7UDR4 H2ZH03 A0A2Y9DDF4 A0A1S3D3V4 A0A2C9K7T3 A0A1B6K2V7 C4B8X4 A0A341CEV0 A0A093R2Y8 S7MG66 F7FU36 C3ZMT5 A0A091JCA6 A0A1W3JEG2 G1SY17 A0A183LJ54 A0A183K760 G1PTD3 U3KPB6 A0A095A255 A0A183RP67 A0A093J9S8 A0A2U3XT83 A0A3Q7SRB7 A0A2F0BCW9 A0A1S3IY04 G4VCB4 A0A087QR25 A0A091WAZ0 A0A2U3V606 A0A1S3I1H4 M3X9Z3 A0A3Q0KMB6 F6UPL6 A0A3M6UUI2 F6Q808 H0UU47 A0A340X040 A0A2Y9FD02 A0A287D6C0 A0A2U3ZDC7 G5AMZ9
A0A154P3P6 A0A0L7KW62 E0W087 D6WQE3 A0A158NCR3 A0A075AJR4 A0A1S8WLA3 A0A151I2R2 A0A310SUH3 A0A183B2V7 A0A3R7GRT6 A0A151XCQ9 E2A4D0 G7Y5T9 A0A151J996 A0A0L7QY67 H2KRA8 A0A1W4WQT0 F4WYR7 A0A3Q0KGB2 A0A1I8HZC7 A0A1I8GZ50 A0A1I8FY27 G4V5J5 A0A074YZD8 A0A088A5U1 A0A195CPE1 A0A183LKS7 A0A2A3EIV7 A0A183NYC3 A0A183K5E4 E4X3B0 T1F7C2 A0A3L8D4R2 A0A1I8INK1 H3BAM5 A0A026W6Y8 A0A095B2K7 A0A0B7B924 E9IM35 A0A0B7B639 A0A0N0BDF6 A0A3S1ASK2 O46178 W4YGB6 V4ABQ0 A0A0B7B9S3 A0A2G8JST0 R7UDR4 H2ZH03 A0A2Y9DDF4 A0A1S3D3V4 A0A2C9K7T3 A0A1B6K2V7 C4B8X4 A0A341CEV0 A0A093R2Y8 S7MG66 F7FU36 C3ZMT5 A0A091JCA6 A0A1W3JEG2 G1SY17 A0A183LJ54 A0A183K760 G1PTD3 U3KPB6 A0A095A255 A0A183RP67 A0A093J9S8 A0A2U3XT83 A0A3Q7SRB7 A0A2F0BCW9 A0A1S3IY04 G4VCB4 A0A087QR25 A0A091WAZ0 A0A2U3V606 A0A1S3I1H4 M3X9Z3 A0A3Q0KMB6 F6UPL6 A0A3M6UUI2 F6Q808 H0UU47 A0A340X040 A0A2Y9FD02 A0A287D6C0 A0A2U3ZDC7 G5AMZ9
Ontologies
PANTHER
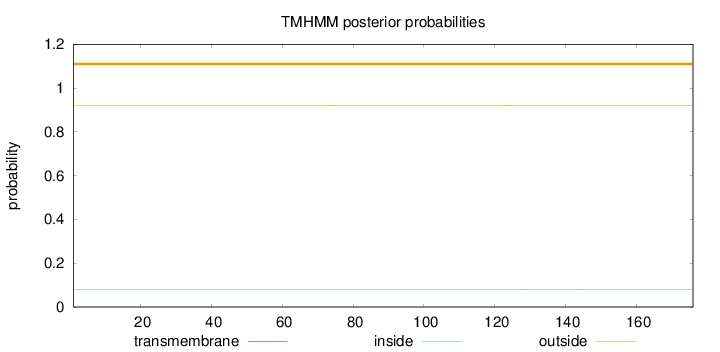
Topology
Length:
176
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00249
Exp number, first 60 AAs:
2e-05
Total prob of N-in:
0.08030
outside
1 - 176
Population Genetic Test Statistics
Pi
290.388966
Theta
181.576377
Tajima's D
2.015551
CLR
0.3539
CSRT
0.883755812209389
Interpretation
Uncertain
