Gene
KWMTBOMO14969
Pre Gene Modal
BGIBMGA000754
Annotation
PREDICTED:_mitochondrial_GTPase_1_[Papilio_machaon]
Full name
Mitochondrial GTPase 1
+ More
Mitochondrial ribosome-associated GTPase 1
Mitochondrial ribosome-associated GTPase 1
Alternative Name
Mitochondrial GTPase 1
GTP-binding protein 7
GTP-binding protein 7
Location in the cell
Mitochondrial Reliability : 2.235
Sequence
CDS
ATGTCCGCTAGATTTGATGGGGCCCTACACAAATTTCGACAGCAATGCCCTTATGTGAGCAAAGACCTGCTCCGCTGGTTCCCTGGTCATATGAACAAAGGTCTAAAACAGATGCAACGTAAATTAAAAAGCGTTGATTGTGTCATAGAAGTACACGATGCCAGGATACCTTTTACGGGCCGAAATCCAATCTTCACAAGCACATTGACTGGCGCTAAACCTCACATATTAGTCCTTAACAAGAGGGATTTAGTCATAACATCATTAATTCCTCGAATTAAAGATCAATTAAAAGCTGAACAAAATGTCGACAATGTCGTGTTTACAAACAGCAAAGACCAGTTTTGCCGCGGTCTGAAGACTATTAAACCTTTAATGGTTGATTTAATAAAAAATTCTAATAGGTATAACAGAAGCGAGGAACTAGAGTATAATGTTATGATAATTGGAGTTCCAAATGTCGGTAAATCCTCTATGATCAACATGTTGAGATCCAGGAATATTAGTGGAAGGCATGTGCTGCCGGTCGGTGCTGTTGCCGGTGTAACCAGGAGTCTTATGATGAAGATGAGAATAAATAATGACCCCTGTATATTTATGTTAGATACTCCAGGTATCTTAGAGCCATCAGTAACAAATATAGAAATGGGTTTGAAGCTTGCATTGTGTGCTGCACTCCAAGATCATCTCGTAGGCGAGGAGATCATCGCAGATTACTTGCTGTACTGGCTTAACAAGCATAGGAAATTTAAATATGTGGACTTCATGGGCTTAGATGAGCCTTGTGATGATATTAATAAGGTACTCCTCTCCGGCGCTATTAAATACAACCGTATCAGAAAAGTTAGAGACTTTGATGGTAAAGTCCGGGATGTACCGGACCTTCTTGAAACCTCCAGACACATTATTAAAGCCTTCAGGACGGGAGAATTGGGAAAAGTCATTCTAGATATTGACCTGTTGGAGAATAGACCTAGCAATAAACAGGAGGTTCTAGCATAA
Protein
MSARFDGALHKFRQQCPYVSKDLLRWFPGHMNKGLKQMQRKLKSVDCVIEVHDARIPFTGRNPIFTSTLTGAKPHILVLNKRDLVITSLIPRIKDQLKAEQNVDNVVFTNSKDQFCRGLKTIKPLMVDLIKNSNRYNRSEELEYNVMIIGVPNVGKSSMINMLRSRNISGRHVLPVGAVAGVTRSLMMKMRINNDPCIFMLDTPGILEPSVTNIEMGLKLALCAALQDHLVGEEIIADYLLYWLNKHRKFKYVDFMGLDEPCDDINKVLLSGAIKYNRIRKVRDFDGKVRDVPDLLETSRHIIKAFRTGELGKVILDIDLLENRPSNKQEVLA
Summary
Description
Plays a role in the regulation of the mitochondrial ribosome assembly and of translational activity. Displays mitochondrial GTPase activity.
Plays a role in the regulation of the mitochondrial ribosome assembly and of translational activity (By similarity). Displays mitochondrial GTPase activity (By similarity).
Plays a role in the regulation of the mitochondrial ribosome assembly and of translational activity (By similarity). Displays mitochondrial GTPase activity (By similarity).
Subunit
Associates with the mitochondrial ribosome large subunit; the association occurs in a GTP-dependent manner (PubMed:23396448).
Similarity
Belongs to the TRAFAC class YlqF/YawG GTPase family. MTG1 subfamily.
Keywords
Complete proteome
GTP-binding
Membrane
Mitochondrion
Mitochondrion inner membrane
Nucleotide-binding
Reference proteome
Transit peptide
Phosphoprotein
Alternative splicing
Polymorphism
Translation regulation
Feature
chain Mitochondrial GTPase 1
splice variant In isoform 2.
sequence variant In dbSNP:rs2255246.
splice variant In isoform 2.
sequence variant In dbSNP:rs2255246.
Uniprot
H9IU24
A0A194RM00
A0A2H1VJ84
A0A194PFF2
A0A2A4JGB3
A0A2W1BJK5
+ More
A0A212EZJ8 A0A3S2NTG8 A0A0L7KVK9 B0XD80 A0A1W5LU01 A0A1Q3F332 A0A1I8PC29 A0A182GJQ3 A0A1I8MNK1 Q17EJ1 J9K0V7 A0A2J7PXP8 A0A067RII7 A0A182R2S2 A0A0L0BNA8 A0A182MIU2 A0A182T2X0 A0A182WAS1 A0A182JVE5 D6WTA4 A0A182YI51 A0A182QDS1 A0A182JJX0 A0A182WZW9 A0A182LI90 A0A182HXG6 A0A182FM84 A0A1A9W1A2 Q7Q2N7 A0A0A1WCH0 A0A182UDV2 A0A182UPB6 A0A2M4CZM8 A0A034W7L8 A0A0K8TRP5 A0A1A9XRH2 A0A182NNA3 A0A0K8UVG2 A0A1A9UFQ3 A0A1J1HLN1 A0A084VV15 A0A1A9Z9Z5 W8BPC4 A0A1B0FIS2 A0A2R7WGF3 A0A0J7KIL4 A0A1B6DW70 A0A336KH73 A0A336MXH6 A0A069DSD3 A0A1Z5L7I4 N6TZ13 A0A1Y1LMD1 A0A195F9R2 A0A0P5ZNI1 E9IM01 A0A154PPH2 A0A0P4VPX9 R4G4B2 A0A1B6LM30 A0A0L7R3K6 A0A2P8ZK25 A0A0M9A1F4 A0A310SQY8 A0A0P4XJ03 A0A3L8D3T5 K7IR66 A0A1B6ET21 T1E815 A0A1B6JAG4 A0A151WXA8 E9FS99 Q29AU5 A0A3B0JLA3 A0A088A7J7 A0A232F132 A0A1W4WF05 A0A2A3E7U7 A0A2M4AZ41 A0A0P4VV26 B4JH48 B4LXB7 E2AK14 B3LZQ5 A0A0P5NR86 B4N9P9 A0A3R7QP68 Q9VCU5 B4R0R1 A0A1W4VMI1 U3DZW3 A0A2R9AGP6 K7DJP7 Q9BT17
A0A212EZJ8 A0A3S2NTG8 A0A0L7KVK9 B0XD80 A0A1W5LU01 A0A1Q3F332 A0A1I8PC29 A0A182GJQ3 A0A1I8MNK1 Q17EJ1 J9K0V7 A0A2J7PXP8 A0A067RII7 A0A182R2S2 A0A0L0BNA8 A0A182MIU2 A0A182T2X0 A0A182WAS1 A0A182JVE5 D6WTA4 A0A182YI51 A0A182QDS1 A0A182JJX0 A0A182WZW9 A0A182LI90 A0A182HXG6 A0A182FM84 A0A1A9W1A2 Q7Q2N7 A0A0A1WCH0 A0A182UDV2 A0A182UPB6 A0A2M4CZM8 A0A034W7L8 A0A0K8TRP5 A0A1A9XRH2 A0A182NNA3 A0A0K8UVG2 A0A1A9UFQ3 A0A1J1HLN1 A0A084VV15 A0A1A9Z9Z5 W8BPC4 A0A1B0FIS2 A0A2R7WGF3 A0A0J7KIL4 A0A1B6DW70 A0A336KH73 A0A336MXH6 A0A069DSD3 A0A1Z5L7I4 N6TZ13 A0A1Y1LMD1 A0A195F9R2 A0A0P5ZNI1 E9IM01 A0A154PPH2 A0A0P4VPX9 R4G4B2 A0A1B6LM30 A0A0L7R3K6 A0A2P8ZK25 A0A0M9A1F4 A0A310SQY8 A0A0P4XJ03 A0A3L8D3T5 K7IR66 A0A1B6ET21 T1E815 A0A1B6JAG4 A0A151WXA8 E9FS99 Q29AU5 A0A3B0JLA3 A0A088A7J7 A0A232F132 A0A1W4WF05 A0A2A3E7U7 A0A2M4AZ41 A0A0P4VV26 B4JH48 B4LXB7 E2AK14 B3LZQ5 A0A0P5NR86 B4N9P9 A0A3R7QP68 Q9VCU5 B4R0R1 A0A1W4VMI1 U3DZW3 A0A2R9AGP6 K7DJP7 Q9BT17
Pubmed
19121390
26354079
28756777
22118469
26227816
26483478
+ More
25315136 17510324 24845553 26108605 18362917 19820115 25244985 20966253 12364791 14747013 17210077 25830018 25348373 26369729 24438588 24495485 26334808 28528879 23537049 28004739 21282665 27129103 29403074 30249741 20075255 21292972 15632085 28648823 17994087 20798317 10731132 12537572 18327897 25243066 22722832 14702039 15164054 15489334 12808030 23396448
25315136 17510324 24845553 26108605 18362917 19820115 25244985 20966253 12364791 14747013 17210077 25830018 25348373 26369729 24438588 24495485 26334808 28528879 23537049 28004739 21282665 27129103 29403074 30249741 20075255 21292972 15632085 28648823 17994087 20798317 10731132 12537572 18327897 25243066 22722832 14702039 15164054 15489334 12808030 23396448
EMBL
BABH01034249
KQ460205
KPJ17011.1
ODYU01002874
SOQ40899.1
KQ459604
+ More
KPI92096.1 NWSH01001601 PCG70748.1 KZ150248 PZC71843.1 AGBW02011278 OWR46910.1 RSAL01000216 RVE44121.1 JTDY01005195 KOB67268.1 DS232745 EDS45334.1 KX034170 ANO54000.1 GFDL01013108 JAV21937.1 JXUM01068660 KQ562512 KXJ75739.1 CH477282 EAT44888.1 ABLF02012332 NEVH01020852 PNF21118.1 KK852455 KDR23631.1 JRES01001623 KNC21398.1 AXCM01000074 KQ971352 EFA06687.1 AXCN02001904 APCN01005301 AAAB01008968 EAA13293.4 GBXI01017633 JAC96658.1 GGFL01006503 MBW70681.1 GAKP01007371 JAC51581.1 GDAI01000564 JAI17039.1 GDHF01021738 JAI30576.1 CVRI01000008 CRK88450.1 ATLV01017144 KE525157 KFB41809.1 GAMC01003565 JAC02991.1 CCAG010023921 KK854789 PTY18747.1 LBMM01007060 KMQ90077.1 GEDC01007386 JAS29912.1 UFQS01000461 UFQT01000461 SSX04214.1 SSX24579.1 UFQS01003326 UFQT01003326 SSX15504.1 SSX34870.1 GBGD01002248 JAC86641.1 GFJQ02003594 JAW03376.1 APGK01045816 KB741039 KB632188 ENN74535.1 ERL89803.1 GEZM01054636 JAV73510.1 KQ981727 KYN36789.1 GDIP01044917 LRGB01000642 JAM58798.1 KZS17218.1 GL764129 EFZ18328.1 KQ435012 KZC13773.1 GDKW01001536 JAI55059.1 ACPB03000196 GAHY01001520 JAA75990.1 GEBQ01015185 JAT24792.1 KQ414663 KOC65414.1 PYGN01000033 PSN56851.1 KQ435789 KOX74436.1 KQ761467 OAD57520.1 GDIP01241963 JAI81438.1 QOIP01000014 RLU15060.1 GECZ01028702 JAS41067.1 GAMD01003148 JAA98442.1 GECU01011530 JAS96176.1 KQ982668 KYQ52510.1 GL732523 EFX89999.1 CM000070 EAL27254.2 OUUW01000007 SPP83005.1 NNAY01001357 OXU24253.1 KZ288347 PBC27584.1 GGFK01012719 MBW46040.1 GDRN01107161 JAI57463.1 CH916369 EDV92739.1 CH940650 EDW67795.1 GL440135 EFN66224.1 CH902617 EDV43049.1 GDIQ01149137 JAL02589.1 CH964232 EDW80614.1 QCYY01002014 ROT73612.1 AE014297 BT022736 AAF56060.2 AAY55152.1 CM000364 EDX13984.1 GAMT01008607 GAMS01007836 GAMR01009917 GAMQ01005381 GAMP01010014 JAB03254.1 JAB15300.1 JAB24015.1 JAB36470.1 JAB42741.1 AJFE02055004 AJFE02055005 AJFE02055006 GABC01001017 GABF01006578 GABD01010043 GABE01002465 JAA10321.1 JAA15567.1 JAA23057.1 JAA42274.1 AK074976 AL161645 AL360181 BC000920 BC004409 BC026039 BC035721
KPI92096.1 NWSH01001601 PCG70748.1 KZ150248 PZC71843.1 AGBW02011278 OWR46910.1 RSAL01000216 RVE44121.1 JTDY01005195 KOB67268.1 DS232745 EDS45334.1 KX034170 ANO54000.1 GFDL01013108 JAV21937.1 JXUM01068660 KQ562512 KXJ75739.1 CH477282 EAT44888.1 ABLF02012332 NEVH01020852 PNF21118.1 KK852455 KDR23631.1 JRES01001623 KNC21398.1 AXCM01000074 KQ971352 EFA06687.1 AXCN02001904 APCN01005301 AAAB01008968 EAA13293.4 GBXI01017633 JAC96658.1 GGFL01006503 MBW70681.1 GAKP01007371 JAC51581.1 GDAI01000564 JAI17039.1 GDHF01021738 JAI30576.1 CVRI01000008 CRK88450.1 ATLV01017144 KE525157 KFB41809.1 GAMC01003565 JAC02991.1 CCAG010023921 KK854789 PTY18747.1 LBMM01007060 KMQ90077.1 GEDC01007386 JAS29912.1 UFQS01000461 UFQT01000461 SSX04214.1 SSX24579.1 UFQS01003326 UFQT01003326 SSX15504.1 SSX34870.1 GBGD01002248 JAC86641.1 GFJQ02003594 JAW03376.1 APGK01045816 KB741039 KB632188 ENN74535.1 ERL89803.1 GEZM01054636 JAV73510.1 KQ981727 KYN36789.1 GDIP01044917 LRGB01000642 JAM58798.1 KZS17218.1 GL764129 EFZ18328.1 KQ435012 KZC13773.1 GDKW01001536 JAI55059.1 ACPB03000196 GAHY01001520 JAA75990.1 GEBQ01015185 JAT24792.1 KQ414663 KOC65414.1 PYGN01000033 PSN56851.1 KQ435789 KOX74436.1 KQ761467 OAD57520.1 GDIP01241963 JAI81438.1 QOIP01000014 RLU15060.1 GECZ01028702 JAS41067.1 GAMD01003148 JAA98442.1 GECU01011530 JAS96176.1 KQ982668 KYQ52510.1 GL732523 EFX89999.1 CM000070 EAL27254.2 OUUW01000007 SPP83005.1 NNAY01001357 OXU24253.1 KZ288347 PBC27584.1 GGFK01012719 MBW46040.1 GDRN01107161 JAI57463.1 CH916369 EDV92739.1 CH940650 EDW67795.1 GL440135 EFN66224.1 CH902617 EDV43049.1 GDIQ01149137 JAL02589.1 CH964232 EDW80614.1 QCYY01002014 ROT73612.1 AE014297 BT022736 AAF56060.2 AAY55152.1 CM000364 EDX13984.1 GAMT01008607 GAMS01007836 GAMR01009917 GAMQ01005381 GAMP01010014 JAB03254.1 JAB15300.1 JAB24015.1 JAB36470.1 JAB42741.1 AJFE02055004 AJFE02055005 AJFE02055006 GABC01001017 GABF01006578 GABD01010043 GABE01002465 JAA10321.1 JAA15567.1 JAA23057.1 JAA42274.1 AK074976 AL161645 AL360181 BC000920 BC004409 BC026039 BC035721
Proteomes
UP000005204
UP000053240
UP000053268
UP000218220
UP000007151
UP000283053
+ More
UP000037510 UP000002320 UP000095300 UP000069940 UP000249989 UP000095301 UP000008820 UP000007819 UP000235965 UP000027135 UP000075900 UP000037069 UP000075883 UP000075901 UP000075920 UP000075881 UP000007266 UP000076408 UP000075886 UP000075880 UP000076407 UP000075882 UP000075840 UP000069272 UP000091820 UP000007062 UP000075902 UP000075903 UP000092443 UP000075884 UP000078200 UP000183832 UP000030765 UP000092445 UP000092444 UP000036403 UP000019118 UP000030742 UP000078541 UP000076858 UP000076502 UP000015103 UP000053825 UP000245037 UP000053105 UP000279307 UP000002358 UP000075809 UP000000305 UP000001819 UP000268350 UP000005203 UP000215335 UP000192223 UP000242457 UP000001070 UP000008792 UP000000311 UP000007801 UP000007798 UP000283509 UP000000803 UP000000304 UP000192221 UP000008225 UP000240080 UP000005640
UP000037510 UP000002320 UP000095300 UP000069940 UP000249989 UP000095301 UP000008820 UP000007819 UP000235965 UP000027135 UP000075900 UP000037069 UP000075883 UP000075901 UP000075920 UP000075881 UP000007266 UP000076408 UP000075886 UP000075880 UP000076407 UP000075882 UP000075840 UP000069272 UP000091820 UP000007062 UP000075902 UP000075903 UP000092443 UP000075884 UP000078200 UP000183832 UP000030765 UP000092445 UP000092444 UP000036403 UP000019118 UP000030742 UP000078541 UP000076858 UP000076502 UP000015103 UP000053825 UP000245037 UP000053105 UP000279307 UP000002358 UP000075809 UP000000305 UP000001819 UP000268350 UP000005203 UP000215335 UP000192223 UP000242457 UP000001070 UP000008792 UP000000311 UP000007801 UP000007798 UP000283509 UP000000803 UP000000304 UP000192221 UP000008225 UP000240080 UP000005640
Interpro
IPR023179
GTP-bd_ortho_bundle_sf
+ More
IPR016478 GTPase_MTG1
IPR027417 P-loop_NTPase
IPR006073 GTP_binding_domain
IPR030378 G_CP_dom
IPR034804 SQR/QFR_C/D
IPR007992 CybS
IPR016130 Tyr_Pase_AS
IPR042165 PTPMT1
IPR029021 Prot-tyrosine_phosphatase-like
IPR000340 Dual-sp_phosphatase_cat-dom
IPR020422 TYR_PHOSPHATASE_DUAL_dom
IPR000387 TYR_PHOSPHATASE_dom
IPR019991 GTP-bd_ribosome_bgen
IPR016478 GTPase_MTG1
IPR027417 P-loop_NTPase
IPR006073 GTP_binding_domain
IPR030378 G_CP_dom
IPR034804 SQR/QFR_C/D
IPR007992 CybS
IPR016130 Tyr_Pase_AS
IPR042165 PTPMT1
IPR029021 Prot-tyrosine_phosphatase-like
IPR000340 Dual-sp_phosphatase_cat-dom
IPR020422 TYR_PHOSPHATASE_DUAL_dom
IPR000387 TYR_PHOSPHATASE_dom
IPR019991 GTP-bd_ribosome_bgen
Gene 3D
ProteinModelPortal
H9IU24
A0A194RM00
A0A2H1VJ84
A0A194PFF2
A0A2A4JGB3
A0A2W1BJK5
+ More
A0A212EZJ8 A0A3S2NTG8 A0A0L7KVK9 B0XD80 A0A1W5LU01 A0A1Q3F332 A0A1I8PC29 A0A182GJQ3 A0A1I8MNK1 Q17EJ1 J9K0V7 A0A2J7PXP8 A0A067RII7 A0A182R2S2 A0A0L0BNA8 A0A182MIU2 A0A182T2X0 A0A182WAS1 A0A182JVE5 D6WTA4 A0A182YI51 A0A182QDS1 A0A182JJX0 A0A182WZW9 A0A182LI90 A0A182HXG6 A0A182FM84 A0A1A9W1A2 Q7Q2N7 A0A0A1WCH0 A0A182UDV2 A0A182UPB6 A0A2M4CZM8 A0A034W7L8 A0A0K8TRP5 A0A1A9XRH2 A0A182NNA3 A0A0K8UVG2 A0A1A9UFQ3 A0A1J1HLN1 A0A084VV15 A0A1A9Z9Z5 W8BPC4 A0A1B0FIS2 A0A2R7WGF3 A0A0J7KIL4 A0A1B6DW70 A0A336KH73 A0A336MXH6 A0A069DSD3 A0A1Z5L7I4 N6TZ13 A0A1Y1LMD1 A0A195F9R2 A0A0P5ZNI1 E9IM01 A0A154PPH2 A0A0P4VPX9 R4G4B2 A0A1B6LM30 A0A0L7R3K6 A0A2P8ZK25 A0A0M9A1F4 A0A310SQY8 A0A0P4XJ03 A0A3L8D3T5 K7IR66 A0A1B6ET21 T1E815 A0A1B6JAG4 A0A151WXA8 E9FS99 Q29AU5 A0A3B0JLA3 A0A088A7J7 A0A232F132 A0A1W4WF05 A0A2A3E7U7 A0A2M4AZ41 A0A0P4VV26 B4JH48 B4LXB7 E2AK14 B3LZQ5 A0A0P5NR86 B4N9P9 A0A3R7QP68 Q9VCU5 B4R0R1 A0A1W4VMI1 U3DZW3 A0A2R9AGP6 K7DJP7 Q9BT17
A0A212EZJ8 A0A3S2NTG8 A0A0L7KVK9 B0XD80 A0A1W5LU01 A0A1Q3F332 A0A1I8PC29 A0A182GJQ3 A0A1I8MNK1 Q17EJ1 J9K0V7 A0A2J7PXP8 A0A067RII7 A0A182R2S2 A0A0L0BNA8 A0A182MIU2 A0A182T2X0 A0A182WAS1 A0A182JVE5 D6WTA4 A0A182YI51 A0A182QDS1 A0A182JJX0 A0A182WZW9 A0A182LI90 A0A182HXG6 A0A182FM84 A0A1A9W1A2 Q7Q2N7 A0A0A1WCH0 A0A182UDV2 A0A182UPB6 A0A2M4CZM8 A0A034W7L8 A0A0K8TRP5 A0A1A9XRH2 A0A182NNA3 A0A0K8UVG2 A0A1A9UFQ3 A0A1J1HLN1 A0A084VV15 A0A1A9Z9Z5 W8BPC4 A0A1B0FIS2 A0A2R7WGF3 A0A0J7KIL4 A0A1B6DW70 A0A336KH73 A0A336MXH6 A0A069DSD3 A0A1Z5L7I4 N6TZ13 A0A1Y1LMD1 A0A195F9R2 A0A0P5ZNI1 E9IM01 A0A154PPH2 A0A0P4VPX9 R4G4B2 A0A1B6LM30 A0A0L7R3K6 A0A2P8ZK25 A0A0M9A1F4 A0A310SQY8 A0A0P4XJ03 A0A3L8D3T5 K7IR66 A0A1B6ET21 T1E815 A0A1B6JAG4 A0A151WXA8 E9FS99 Q29AU5 A0A3B0JLA3 A0A088A7J7 A0A232F132 A0A1W4WF05 A0A2A3E7U7 A0A2M4AZ41 A0A0P4VV26 B4JH48 B4LXB7 E2AK14 B3LZQ5 A0A0P5NR86 B4N9P9 A0A3R7QP68 Q9VCU5 B4R0R1 A0A1W4VMI1 U3DZW3 A0A2R9AGP6 K7DJP7 Q9BT17
PDB
1PUJ
E-value=1.23494e-26,
Score=297
Ontologies
GO
PANTHER
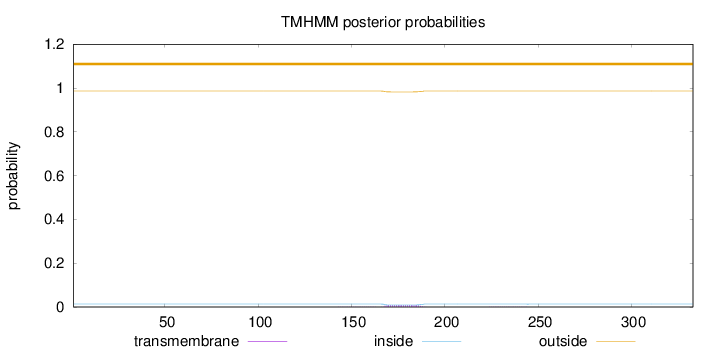
Topology
Subcellular location
Mitochondrion inner membrane
Length:
333
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.18546
Exp number, first 60 AAs:
3e-05
Total prob of N-in:
0.01377
outside
1 - 333
Population Genetic Test Statistics
Pi
218.282125
Theta
163.259613
Tajima's D
0.610863
CLR
0.314404
CSRT
0.538873056347183
Interpretation
Uncertain
