Gene
KWMTBOMO14967 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA000819
Annotation
hypothetical_protein_KGM_16514_[Danaus_plexippus]
Location in the cell
Cytoplasmic Reliability : 2.903
Sequence
CDS
ATGAAGAAGGTCGTGATTTTTGGTTCGACTGGCGTGATCGGCTTGAACGCTGTTGAGGCGGCACTTAAAAAAGGCCTAGAAGTACGAGCGTTCGTACGTGACCCAGCAAAACTCCCAGAACATCTTAAAGACAAGGTCGAGATAGTCAAAGGAAATGTTCTCGAACCAGACTCGGTACATGAGGCCGTCGAAGGAACAGACGCTGTCGTCATCACCCTAGGGACTAGGAACGACCTGGCGCCGACTTCGGATTTATCTGAAGGAACCAAAAACATAATAGACGCGATGAGAGCGAAGAACGTTAAGACGGTGTCGGCTTGTTTATCGGCGTTCTTATTTTATGAACAAGAAAAGGTGCCGCCGATCTTCGTCAATTTGAACGAAGACCACAAGAGAATGTTCCAGGCTCTGAAAGACAGCGGCTTAAATTGGATCGCCGCGTTTCCGCCACACTTCACAGACGACCCAAGCCGAGAAATGATTATTGAAGTGAACCCTGAGAAGACACCGGGCAGGACCATTGCCAAGTGCGACCTCGGCACATTCCTAGTGGACGCGCTTTCCGAACCCAAGTACTACAAGGCAGTCATTGGCATCTGCAATGTGCCCAAAGAATGA
Protein
MKKVVIFGSTGVIGLNAVEAALKKGLEVRAFVRDPAKLPEHLKDKVEIVKGNVLEPDSVHEAVEGTDAVVITLGTRNDLAPTSDLSEGTKNIIDAMRAKNVKTVSACLSAFLFYEQEKVPPIFVNLNEDHKRMFQALKDSGLNWIAAFPPHFTDDPSREMIIEVNPEKTPGRTIAKCDLGTFLVDALSEPKYYKAVIGICNVPKE
Summary
Uniprot
H9IU89
A0A2P1AG33
A0A3S2NCR9
S4NN31
A0A212EZI4
A0A0L7L7A5
+ More
A0A2A4JH48 A0A2W1BD89 A0A194PFN4 A0A194RGR9 A0A2H1VJD1 A0A2P1AG40 A0A1W4WYK6 D6WR30 A0A154PHB6 A0A0L7R1G1 A0A2S2R3Q5 A0A0M8ZSU3 A0A2S2PQX4 N6UCT5 A0A1Y1JR15 A0A026W9R0 A0A2P8XNU9 A0A2A3ERP7 C4WRU5 A0A088A0I0 K7IQD9 A0A232F2Z7 A0A151XBW5 V5GS29 A0A195CRH6 A0A0T6B4D1 A0A195EFE4 A0A2J7QCY9 A0A336L5M3 E9IBH1 A0A0C9PTT5 A0A336KSN1 U5EU66 A0A1B6KV41 A0A195BG41 A0A158NTG4 E2AI27 A0A0P4WE64 Q16FC8 A0A1B6JSL3 A0A1B6FYX7 A0A1L8DFY0 A0A1B6HFB9 A0A1L8DFM8 N6UBY7 A0A1B6K8U4 A0A1B6L9I4 E0VYG9 A0A1B6EI75 A0A1B6FNU8 A0A182JAH9 A0A067RAP8 A0A1B6HAD4 A0A1B6L7J4 A0A2M4AWR7 A0A0P6GDC1 A0A1B0CGQ9 A0A1J1I6S2 A0A1Y9J125 A0A2M4AX84 A0A3F2YUS3 A0A182U5L7 A0A2C9GQH8 A0A182ULT2 Q7PXB6 W5J782 F4WG16 V9LDT3 T1DGH2 A0A151JWJ0 B7PJY6 V9LCH4 A0A182Q825 V5HZ16 B0WX84 E9IBH0 A0A131Y0E3 A0A182RNK9 T1E3U4 A0A3R7MQQ2 A0A0K8TQW4 V5IC73 A0A0E9X3H4 T1JSZ1 A0A3P8ZZZ5 A0A182M9R6 A0A1A7ZEI1 A0A3P9DE68 A0A3P8QCG9 A0A087UVC3 A0A3Q2UUG1 A0A182YBC7 A0A1A8IND5 V9ICV4
A0A2A4JH48 A0A2W1BD89 A0A194PFN4 A0A194RGR9 A0A2H1VJD1 A0A2P1AG40 A0A1W4WYK6 D6WR30 A0A154PHB6 A0A0L7R1G1 A0A2S2R3Q5 A0A0M8ZSU3 A0A2S2PQX4 N6UCT5 A0A1Y1JR15 A0A026W9R0 A0A2P8XNU9 A0A2A3ERP7 C4WRU5 A0A088A0I0 K7IQD9 A0A232F2Z7 A0A151XBW5 V5GS29 A0A195CRH6 A0A0T6B4D1 A0A195EFE4 A0A2J7QCY9 A0A336L5M3 E9IBH1 A0A0C9PTT5 A0A336KSN1 U5EU66 A0A1B6KV41 A0A195BG41 A0A158NTG4 E2AI27 A0A0P4WE64 Q16FC8 A0A1B6JSL3 A0A1B6FYX7 A0A1L8DFY0 A0A1B6HFB9 A0A1L8DFM8 N6UBY7 A0A1B6K8U4 A0A1B6L9I4 E0VYG9 A0A1B6EI75 A0A1B6FNU8 A0A182JAH9 A0A067RAP8 A0A1B6HAD4 A0A1B6L7J4 A0A2M4AWR7 A0A0P6GDC1 A0A1B0CGQ9 A0A1J1I6S2 A0A1Y9J125 A0A2M4AX84 A0A3F2YUS3 A0A182U5L7 A0A2C9GQH8 A0A182ULT2 Q7PXB6 W5J782 F4WG16 V9LDT3 T1DGH2 A0A151JWJ0 B7PJY6 V9LCH4 A0A182Q825 V5HZ16 B0WX84 E9IBH0 A0A131Y0E3 A0A182RNK9 T1E3U4 A0A3R7MQQ2 A0A0K8TQW4 V5IC73 A0A0E9X3H4 T1JSZ1 A0A3P8ZZZ5 A0A182M9R6 A0A1A7ZEI1 A0A3P9DE68 A0A3P8QCG9 A0A087UVC3 A0A3Q2UUG1 A0A182YBC7 A0A1A8IND5 V9ICV4
Pubmed
19121390
23622113
22118469
26227816
28756777
26354079
+ More
18362917 19820115 23537049 28004739 24508170 30249741 29403074 20075255 28648823 21282665 21347285 20798317 17510324 20566863 24845553 20966253 12364791 14747013 17210077 20920257 23761445 21719571 24402279 25765539 24330624 26369729 25613341 25069045 25186727 25244985
18362917 19820115 23537049 28004739 24508170 30249741 29403074 20075255 28648823 21282665 21347285 20798317 17510324 20566863 24845553 20966253 12364791 14747013 17210077 20920257 23761445 21719571 24402279 25765539 24330624 26369729 25613341 25069045 25186727 25244985
EMBL
BABH01034249
KY825252
AVI24059.1
RSAL01000216
RVE44120.1
GAIX01014111
+ More
JAA78449.1 AGBW02011278 OWR46908.1 JTDY01002537 KOB71224.1 NWSH01001601 PCG70750.1 KZ150248 PZC71845.1 KQ459604 KPI92097.1 KQ460205 KPJ17013.1 ODYU01002874 SOQ40901.1 KY825254 AVI24061.1 KQ971354 EFA07011.1 KQ434905 KZC11233.1 KQ414667 KOC64684.1 GGMS01015443 MBY84646.1 KQ435863 KOX70420.1 GGMR01019240 MBY31859.1 APGK01028661 KB740694 ENN79480.1 GEZM01102985 GEZM01102984 GEZM01102983 GEZM01102982 GEZM01102981 GEZM01102980 JAV51649.1 KK107341 QOIP01000012 EZA52406.1 RLU16150.1 PYGN01001629 PSN33680.1 KZ288194 PBC33886.1 ABLF02026465 AK339918 BAH70615.1 NNAY01001084 OXU25206.1 KQ982320 KYQ57788.1 GALX01005423 JAB63043.1 KQ977381 KYN03097.1 LJIG01009912 KRT82132.1 KQ978957 KYN26968.1 NEVH01015852 PNF26446.1 UFQS01001683 UFQT01001683 SSX11849.1 SSX31413.1 GL762111 EFZ22031.1 GBYB01004743 GBYB01004748 JAG74510.1 JAG74515.1 UFQS01000834 UFQT01000834 SSX07148.1 SSX27491.1 GANO01002456 JAB57415.1 GEBQ01024672 JAT15305.1 KQ976488 KYM83580.1 ADTU01025892 ADTU01025893 GL439621 EFN66935.1 GDRN01077515 GDRN01077514 GDRN01077513 JAI62725.1 CH478462 EAT32941.1 GECU01005522 JAT02185.1 GECZ01014931 GECZ01014385 GECZ01010414 GECZ01010319 JAS54838.1 JAS55384.1 JAS59355.1 JAS59450.1 GFDF01008799 JAV05285.1 GECU01034395 JAS73311.1 GFDF01008825 JAV05259.1 APGK01035014 KB740923 KB632326 ENN78146.1 ERL92430.1 GEBQ01032117 JAT07860.1 GEBQ01019619 JAT20358.1 DS235845 EEB18425.1 GECZ01032134 JAS37635.1 GECZ01017884 GECZ01016815 JAS51885.1 JAS52954.1 KK852577 KDR20925.1 GECU01036072 JAS71634.1 GEBQ01025458 GEBQ01020337 GEBQ01017156 GEBQ01015899 JAT14519.1 JAT19640.1 JAT22821.1 JAT24078.1 GGFK01011918 MBW45239.1 GDIP01043157 GDIQ01034931 LRGB01000781 JAM60558.1 JAN59806.1 KZS15994.1 AJWK01011552 CVRI01000039 CRK94646.1 GGFK01011887 MBW45208.1 APCN01000836 AAAB01008987 EAA01772.3 ADMH02002125 ETN58725.1 GL888128 EGI66783.1 JW877741 AFP10258.1 GAMD01002776 JAA98814.1 KQ981639 KYN38690.1 ABJB010101752 ABJB010124304 ABJB010152675 ABJB010824106 ABJB010940102 ABJB010960117 ABJB011106913 ABJB011128215 DS729150 EEC06908.1 JW877581 AFP10098.1 AXCN02001519 GANP01000344 JAB84124.1 DS232159 EDS36345.1 EFZ22041.1 GEFM01003067 JAP72729.1 GALA01000055 JAA94797.1 QCYY01000688 ROT83404.1 GDAI01000854 JAI16749.1 GANP01014187 JAB70281.1 GBXM01011546 JAH97031.1 CAEY01000468 AXCM01000656 HADY01002000 HAEJ01017224 SBP40485.1 KK121832 KFM81312.1 HAED01012380 HAEE01001528 SBQ98774.1 JR038132 AEY58124.1
JAA78449.1 AGBW02011278 OWR46908.1 JTDY01002537 KOB71224.1 NWSH01001601 PCG70750.1 KZ150248 PZC71845.1 KQ459604 KPI92097.1 KQ460205 KPJ17013.1 ODYU01002874 SOQ40901.1 KY825254 AVI24061.1 KQ971354 EFA07011.1 KQ434905 KZC11233.1 KQ414667 KOC64684.1 GGMS01015443 MBY84646.1 KQ435863 KOX70420.1 GGMR01019240 MBY31859.1 APGK01028661 KB740694 ENN79480.1 GEZM01102985 GEZM01102984 GEZM01102983 GEZM01102982 GEZM01102981 GEZM01102980 JAV51649.1 KK107341 QOIP01000012 EZA52406.1 RLU16150.1 PYGN01001629 PSN33680.1 KZ288194 PBC33886.1 ABLF02026465 AK339918 BAH70615.1 NNAY01001084 OXU25206.1 KQ982320 KYQ57788.1 GALX01005423 JAB63043.1 KQ977381 KYN03097.1 LJIG01009912 KRT82132.1 KQ978957 KYN26968.1 NEVH01015852 PNF26446.1 UFQS01001683 UFQT01001683 SSX11849.1 SSX31413.1 GL762111 EFZ22031.1 GBYB01004743 GBYB01004748 JAG74510.1 JAG74515.1 UFQS01000834 UFQT01000834 SSX07148.1 SSX27491.1 GANO01002456 JAB57415.1 GEBQ01024672 JAT15305.1 KQ976488 KYM83580.1 ADTU01025892 ADTU01025893 GL439621 EFN66935.1 GDRN01077515 GDRN01077514 GDRN01077513 JAI62725.1 CH478462 EAT32941.1 GECU01005522 JAT02185.1 GECZ01014931 GECZ01014385 GECZ01010414 GECZ01010319 JAS54838.1 JAS55384.1 JAS59355.1 JAS59450.1 GFDF01008799 JAV05285.1 GECU01034395 JAS73311.1 GFDF01008825 JAV05259.1 APGK01035014 KB740923 KB632326 ENN78146.1 ERL92430.1 GEBQ01032117 JAT07860.1 GEBQ01019619 JAT20358.1 DS235845 EEB18425.1 GECZ01032134 JAS37635.1 GECZ01017884 GECZ01016815 JAS51885.1 JAS52954.1 KK852577 KDR20925.1 GECU01036072 JAS71634.1 GEBQ01025458 GEBQ01020337 GEBQ01017156 GEBQ01015899 JAT14519.1 JAT19640.1 JAT22821.1 JAT24078.1 GGFK01011918 MBW45239.1 GDIP01043157 GDIQ01034931 LRGB01000781 JAM60558.1 JAN59806.1 KZS15994.1 AJWK01011552 CVRI01000039 CRK94646.1 GGFK01011887 MBW45208.1 APCN01000836 AAAB01008987 EAA01772.3 ADMH02002125 ETN58725.1 GL888128 EGI66783.1 JW877741 AFP10258.1 GAMD01002776 JAA98814.1 KQ981639 KYN38690.1 ABJB010101752 ABJB010124304 ABJB010152675 ABJB010824106 ABJB010940102 ABJB010960117 ABJB011106913 ABJB011128215 DS729150 EEC06908.1 JW877581 AFP10098.1 AXCN02001519 GANP01000344 JAB84124.1 DS232159 EDS36345.1 EFZ22041.1 GEFM01003067 JAP72729.1 GALA01000055 JAA94797.1 QCYY01000688 ROT83404.1 GDAI01000854 JAI16749.1 GANP01014187 JAB70281.1 GBXM01011546 JAH97031.1 CAEY01000468 AXCM01000656 HADY01002000 HAEJ01017224 SBP40485.1 KK121832 KFM81312.1 HAED01012380 HAEE01001528 SBQ98774.1 JR038132 AEY58124.1
Proteomes
UP000005204
UP000283053
UP000007151
UP000037510
UP000218220
UP000053268
+ More
UP000053240 UP000192223 UP000007266 UP000076502 UP000053825 UP000053105 UP000019118 UP000053097 UP000279307 UP000245037 UP000242457 UP000007819 UP000005203 UP000002358 UP000215335 UP000075809 UP000078542 UP000078492 UP000235965 UP000078540 UP000005205 UP000000311 UP000008820 UP000030742 UP000009046 UP000075880 UP000027135 UP000076858 UP000092461 UP000183832 UP000076407 UP000075882 UP000075902 UP000075840 UP000075903 UP000007062 UP000000673 UP000007755 UP000078541 UP000001555 UP000075886 UP000002320 UP000075900 UP000283509 UP000015104 UP000265140 UP000075883 UP000265160 UP000265100 UP000054359 UP000264840 UP000076408
UP000053240 UP000192223 UP000007266 UP000076502 UP000053825 UP000053105 UP000019118 UP000053097 UP000279307 UP000245037 UP000242457 UP000007819 UP000005203 UP000002358 UP000215335 UP000075809 UP000078542 UP000078492 UP000235965 UP000078540 UP000005205 UP000000311 UP000008820 UP000030742 UP000009046 UP000075880 UP000027135 UP000076858 UP000092461 UP000183832 UP000076407 UP000075882 UP000075902 UP000075840 UP000075903 UP000007062 UP000000673 UP000007755 UP000078541 UP000001555 UP000075886 UP000002320 UP000075900 UP000283509 UP000015104 UP000265140 UP000075883 UP000265160 UP000265100 UP000054359 UP000264840 UP000076408
Interpro
SUPFAM
SSF51735
SSF51735
Gene 3D
ProteinModelPortal
H9IU89
A0A2P1AG33
A0A3S2NCR9
S4NN31
A0A212EZI4
A0A0L7L7A5
+ More
A0A2A4JH48 A0A2W1BD89 A0A194PFN4 A0A194RGR9 A0A2H1VJD1 A0A2P1AG40 A0A1W4WYK6 D6WR30 A0A154PHB6 A0A0L7R1G1 A0A2S2R3Q5 A0A0M8ZSU3 A0A2S2PQX4 N6UCT5 A0A1Y1JR15 A0A026W9R0 A0A2P8XNU9 A0A2A3ERP7 C4WRU5 A0A088A0I0 K7IQD9 A0A232F2Z7 A0A151XBW5 V5GS29 A0A195CRH6 A0A0T6B4D1 A0A195EFE4 A0A2J7QCY9 A0A336L5M3 E9IBH1 A0A0C9PTT5 A0A336KSN1 U5EU66 A0A1B6KV41 A0A195BG41 A0A158NTG4 E2AI27 A0A0P4WE64 Q16FC8 A0A1B6JSL3 A0A1B6FYX7 A0A1L8DFY0 A0A1B6HFB9 A0A1L8DFM8 N6UBY7 A0A1B6K8U4 A0A1B6L9I4 E0VYG9 A0A1B6EI75 A0A1B6FNU8 A0A182JAH9 A0A067RAP8 A0A1B6HAD4 A0A1B6L7J4 A0A2M4AWR7 A0A0P6GDC1 A0A1B0CGQ9 A0A1J1I6S2 A0A1Y9J125 A0A2M4AX84 A0A3F2YUS3 A0A182U5L7 A0A2C9GQH8 A0A182ULT2 Q7PXB6 W5J782 F4WG16 V9LDT3 T1DGH2 A0A151JWJ0 B7PJY6 V9LCH4 A0A182Q825 V5HZ16 B0WX84 E9IBH0 A0A131Y0E3 A0A182RNK9 T1E3U4 A0A3R7MQQ2 A0A0K8TQW4 V5IC73 A0A0E9X3H4 T1JSZ1 A0A3P8ZZZ5 A0A182M9R6 A0A1A7ZEI1 A0A3P9DE68 A0A3P8QCG9 A0A087UVC3 A0A3Q2UUG1 A0A182YBC7 A0A1A8IND5 V9ICV4
A0A2A4JH48 A0A2W1BD89 A0A194PFN4 A0A194RGR9 A0A2H1VJD1 A0A2P1AG40 A0A1W4WYK6 D6WR30 A0A154PHB6 A0A0L7R1G1 A0A2S2R3Q5 A0A0M8ZSU3 A0A2S2PQX4 N6UCT5 A0A1Y1JR15 A0A026W9R0 A0A2P8XNU9 A0A2A3ERP7 C4WRU5 A0A088A0I0 K7IQD9 A0A232F2Z7 A0A151XBW5 V5GS29 A0A195CRH6 A0A0T6B4D1 A0A195EFE4 A0A2J7QCY9 A0A336L5M3 E9IBH1 A0A0C9PTT5 A0A336KSN1 U5EU66 A0A1B6KV41 A0A195BG41 A0A158NTG4 E2AI27 A0A0P4WE64 Q16FC8 A0A1B6JSL3 A0A1B6FYX7 A0A1L8DFY0 A0A1B6HFB9 A0A1L8DFM8 N6UBY7 A0A1B6K8U4 A0A1B6L9I4 E0VYG9 A0A1B6EI75 A0A1B6FNU8 A0A182JAH9 A0A067RAP8 A0A1B6HAD4 A0A1B6L7J4 A0A2M4AWR7 A0A0P6GDC1 A0A1B0CGQ9 A0A1J1I6S2 A0A1Y9J125 A0A2M4AX84 A0A3F2YUS3 A0A182U5L7 A0A2C9GQH8 A0A182ULT2 Q7PXB6 W5J782 F4WG16 V9LDT3 T1DGH2 A0A151JWJ0 B7PJY6 V9LCH4 A0A182Q825 V5HZ16 B0WX84 E9IBH0 A0A131Y0E3 A0A182RNK9 T1E3U4 A0A3R7MQQ2 A0A0K8TQW4 V5IC73 A0A0E9X3H4 T1JSZ1 A0A3P8ZZZ5 A0A182M9R6 A0A1A7ZEI1 A0A3P9DE68 A0A3P8QCG9 A0A087UVC3 A0A3Q2UUG1 A0A182YBC7 A0A1A8IND5 V9ICV4
PDB
5OOH
E-value=2.66493e-40,
Score=412
Ontologies
PATHWAY
PANTHER
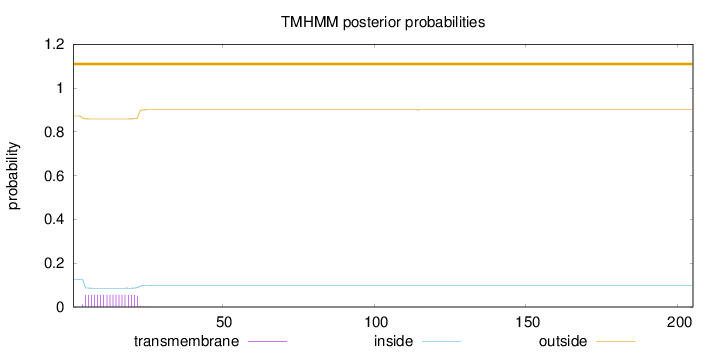
Topology
Length:
205
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.01176
Exp number, first 60 AAs:
1.0078
Total prob of N-in:
0.12722
outside
1 - 205
Population Genetic Test Statistics
Pi
297.230922
Theta
163.815266
Tajima's D
2.414756
CLR
0.334262
CSRT
0.936903154842258
Interpretation
Uncertain
