Pre Gene Modal
BGIBMGA000758
Annotation
PREDICTED:_protein-lysine_N-methyltransferase_N6amt2_[Amyelois_transitella]
Full name
Protein-lysine N-methyltransferase 101735505
+ More
Protein-lysine N-methyltransferase SFRICE_009493
Protein-lysine N-methyltransferase B5V51_820
Protein-lysine N-methyltransferase B5X24_HaOG212382
Protein-lysine N-methyltransferase
Protein-lysine N-methyltransferase RR48_13872
Protein-lysine N-methyltransferase KGM_212814
Protein-lysine N-methyltransferase evm_011229
Protein-lysine N-methyltransferase TcasGA2_TC034616
Protein-lysine N-methyltransferase TSAR_004185
Protein-lysine N-methyltransferase RF55_3503
Protein-lysine N-methyltransferase EAG_03757
Protein-lysine N-methyltransferase WH47_12669
Protein-lysine N-methyltransferase LOC108734122
Protein-lysine N-methyltransferase g.8434
Protein-lysine N-methyltransferase EAI_14662
Protein-lysine N-methyltransferase WN48_04004
Protein-lysine N-methyltransferase DAPPUDRAFT_52203
Protein-lysine N-methyltransferase APZ42_019783
Protein-lysine N-methyltransferase L798_06665
EEF1A lysine methyltransferase 1
Protein-lysine N-methyltransferase B7P43_G04479
Protein-lysine N-methyltransferase C0J52_16608
Protein-lysine N-methyltransferase D910_00984
Protein-lysine N-methyltransferase WN55_03718
Protein-lysine N-methyltransferase APICC_00557
Protein-lysine N-methyltransferase G5I_04406
Protein-lysine N-methyltransferase n6amt2
Protein-lysine N-methyltransferase LOC552772
Protein-lysine N-methyltransferase ALC53_12830
Protein-lysine N-methyltransferase ACYPI004839
Protein-lysine N-methyltransferase 105623729
Protein-lysine N-methyltransferase SFRICE_009493
Protein-lysine N-methyltransferase B5V51_820
Protein-lysine N-methyltransferase B5X24_HaOG212382
Protein-lysine N-methyltransferase
Protein-lysine N-methyltransferase RR48_13872
Protein-lysine N-methyltransferase KGM_212814
Protein-lysine N-methyltransferase evm_011229
Protein-lysine N-methyltransferase TcasGA2_TC034616
Protein-lysine N-methyltransferase TSAR_004185
Protein-lysine N-methyltransferase RF55_3503
Protein-lysine N-methyltransferase EAG_03757
Protein-lysine N-methyltransferase WH47_12669
Protein-lysine N-methyltransferase LOC108734122
Protein-lysine N-methyltransferase g.8434
Protein-lysine N-methyltransferase EAI_14662
Protein-lysine N-methyltransferase WN48_04004
Protein-lysine N-methyltransferase DAPPUDRAFT_52203
Protein-lysine N-methyltransferase APZ42_019783
Protein-lysine N-methyltransferase L798_06665
EEF1A lysine methyltransferase 1
Protein-lysine N-methyltransferase B7P43_G04479
Protein-lysine N-methyltransferase C0J52_16608
Protein-lysine N-methyltransferase D910_00984
Protein-lysine N-methyltransferase WN55_03718
Protein-lysine N-methyltransferase APICC_00557
Protein-lysine N-methyltransferase G5I_04406
Protein-lysine N-methyltransferase n6amt2
Protein-lysine N-methyltransferase LOC552772
Protein-lysine N-methyltransferase ALC53_12830
Protein-lysine N-methyltransferase ACYPI004839
Protein-lysine N-methyltransferase 105623729
Alternative Name
N(6)-adenine-specific DNA methyltransferase 2
Protein-lysine N-methyltransferase N6AMT2
Protein-lysine N-methyltransferase N6amt2
Protein-lysine N-methyltransferase n6amt2
Protein-lysine N-methyltransferase N6AMT2
Protein-lysine N-methyltransferase N6amt2
Protein-lysine N-methyltransferase n6amt2
Location in the cell
Cytoplasmic Reliability : 2.524
Sequence
CDS
ATGGAAGCAGATGAAGACGTGCCCACATTGTCAGCAGAGACATTTGCTGCTTTACAGGAATTCTATGCTGAACAAAGTAAAAGACAAGAAATCCTTGTAAAGCTAGAAGCGGATAAAAAATTAACTGAGAATATTTTGTTTGATGAAAATTGGCAACTGAGTCAGTTTTGGTATGATGAGAAAACGGTACACTCTCTGGTCAAAGTAATTGATAAAGTATTAGACGACAGAGGTAAAGTGGCACTGATTTCGTGCCCCACACTCTTTGTGCCCTTGAAAAGACAGATCGGAGACAGAGGGACTGTCACCCTACTAGAATACGACAGACGATTTGAAGTTCACGGTCCTGACTACATATTCTACGATTATAACAATCCTAAGGAGGTACCACCTGACGTACATCACTCATACGATTTAGTAGTGGCCGACCCACCATTCCTTTCTGAAGAATGCATAACAAAAACATCGGAGACAATCAAATTGCTTTCGAAAGATAAAATAATTTTATGCACCGGTACCATAATGAAGGATATAGTAAAGGAATTGCTAGATTTAAAATTATGCGAATTCCAACCTAAACACCGAAATAATTTAGCTAATGAGTTCTCGTGTTATGCTAACTTTGATTTGGACAGTGTCTTGAGTTGA
Protein
MEADEDVPTLSAETFAALQEFYAEQSKRQEILVKLEADKKLTENILFDENWQLSQFWYDEKTVHSLVKVIDKVLDDRGKVALISCPTLFVPLKRQIGDRGTVTLLEYDRRFEVHGPDYIFYDYNNPKEVPPDVHHSYDLVVADPPFLSEECITKTSETIKLLSKDKIILCTGTIMKDIVKELLDLKLCEFQPKHRNNLANEFSCYANFDLDSVLS
Summary
Description
S-adenosyl-L-methionine-dependent protein-lysine N-methyltransferase that methylates elongation factor 1-alpha.
Protein-lysine methyltransferase that selectively catalyzes the trimethylation of EEF1A at 'Lys-79'.
Protein-lysine methyltransferase that selectively catalyzes the trimethylation of EEF1A at 'Lys-79'.
Similarity
Belongs to the class I-like SAM-binding methyltransferase superfamily. EFM5 family.
Keywords
Acetylation
Complete proteome
Cytoplasm
Methyltransferase
Phosphoprotein
Reference proteome
Transferase
Feature
chain EEF1A lysine methyltransferase 1
Uniprot
H9IU28
A0A2H1VX90
A0A2A4JLY0
A0A2W1BGC8
I4DP87
A0A194RM05
+ More
A0A212EZH5 S4PB98 A0A3S2NN80 A0A067ZWL4 D7EJW9 A0A232FH62 A0A0J7L0V8 E2B162 A0A0L7QKN8 A0A1W4WAJ5 K7J0S4 A0A0V0GD98 A0A1B6KMJ6 A0A0N7Z9H2 R4G8M8 A0A023F731 A0A1B6GV00 E2C1V5 A0A0P4WSG7 A0A0P5SJM6 A0A310SAI0 E9GLC9 A0A0P5T5A5 A0A0P5SJS5 A0A0P5E3W9 E9JD83 A0A067RI41 A0A0P5T1M3 A0A0P5AAG4 C1C3V5 A0A2J7QW40 A0A0P6JKW8 A0A2P8Y4X4 A0A093KAE6 G1KAT8 U4URX5 A0A218UFW3 A0A1X7UYL4 H1A020 A0A0P5A052 A0A096MQR7 A0A0D9RZV1 A0A2K6NLP9 A0A2K5KVP5 N6T7F1 G7PVT6 G7NJQ2 A0A154PL59 A0A2K6DTB8 A0A2K5VXZ2 I2CY46 H0ZM64 A0A2K5XE51 A0A2K5I4G4 H0XTS6 A0A2A3EA95 F4WFJ7 A0A0P5ECT1 Q9CY45 A0A091EJ22 B7ZUJ8 A9JSB0 A0A091GPC2 A0A2K6MVW9 A0A0D2X7Z2 U3K265 A0A2H8TDH0 A0A2K6EJ59 A0A1A6G1X7 A0A087ZRC4 A0A3Q0G2W6 A0A1U7ULI8 A0A099YVG5 A0A1L8HAR5 H0VJ85 A0A093GSI4 A0A091NB18 A0A2R9C8B4 H2Q794 A0A087R290 F7EL08 A0A151HYN4 G3TAT6 C4WUN5 L9JBD9 A0A093F974 A0A091FV71 B5DEG5 A0A091PA60 G3RL82 Q6GN98 A0A158NSJ1 I3MFY8 K7AG74
A0A212EZH5 S4PB98 A0A3S2NN80 A0A067ZWL4 D7EJW9 A0A232FH62 A0A0J7L0V8 E2B162 A0A0L7QKN8 A0A1W4WAJ5 K7J0S4 A0A0V0GD98 A0A1B6KMJ6 A0A0N7Z9H2 R4G8M8 A0A023F731 A0A1B6GV00 E2C1V5 A0A0P4WSG7 A0A0P5SJM6 A0A310SAI0 E9GLC9 A0A0P5T5A5 A0A0P5SJS5 A0A0P5E3W9 E9JD83 A0A067RI41 A0A0P5T1M3 A0A0P5AAG4 C1C3V5 A0A2J7QW40 A0A0P6JKW8 A0A2P8Y4X4 A0A093KAE6 G1KAT8 U4URX5 A0A218UFW3 A0A1X7UYL4 H1A020 A0A0P5A052 A0A096MQR7 A0A0D9RZV1 A0A2K6NLP9 A0A2K5KVP5 N6T7F1 G7PVT6 G7NJQ2 A0A154PL59 A0A2K6DTB8 A0A2K5VXZ2 I2CY46 H0ZM64 A0A2K5XE51 A0A2K5I4G4 H0XTS6 A0A2A3EA95 F4WFJ7 A0A0P5ECT1 Q9CY45 A0A091EJ22 B7ZUJ8 A9JSB0 A0A091GPC2 A0A2K6MVW9 A0A0D2X7Z2 U3K265 A0A2H8TDH0 A0A2K6EJ59 A0A1A6G1X7 A0A087ZRC4 A0A3Q0G2W6 A0A1U7ULI8 A0A099YVG5 A0A1L8HAR5 H0VJ85 A0A093GSI4 A0A091NB18 A0A2R9C8B4 H2Q794 A0A087R290 F7EL08 A0A151HYN4 G3TAT6 C4WUN5 L9JBD9 A0A093F974 A0A091FV71 B5DEG5 A0A091PA60 G3RL82 Q6GN98 A0A158NSJ1 I3MFY8 K7AG74
EC Number
2.1.1.-
Pubmed
19121390
28756777
22651552
26354079
22118469
23622113
+ More
18362917 19820115 28648823 20798317 20075255 27129103 25474469 21292972 21282665 24845553 29403074 23537049 20360741 25362486 22002653 17431167 25319552 21719571 16141072 15489334 21183079 20431018 12040188 19468303 27762356 21993624 22722832 16136131 18464734 23385571 15057822 21347285
18362917 19820115 28648823 20798317 20075255 27129103 25474469 21292972 21282665 24845553 29403074 23537049 20360741 25362486 22002653 17431167 25319552 21719571 16141072 15489334 21183079 20431018 12040188 19468303 27762356 21993624 22722832 16136131 18464734 23385571 15057822 21347285
EMBL
BABH01034246
ODYU01005008
SOQ45467.1
NWSH01001137
PCG72443.1
KZ150248
+ More
PZC71850.1 AK403471 BAM19727.1 KQ460205 KPJ17016.1 AGBW02011278 OWR46905.1 GAIX01006022 JAA86538.1 RSAL01000216 RVE44118.1 KF294266 AHZ08394.1 KQ971327 KYB28399.1 NNAY01000195 OXU30096.1 LBMM01001488 KMQ96223.1 GL444789 EFN60586.1 KQ414954 KOC59076.1 GECL01000759 JAP05365.1 GEBQ01027320 JAT12657.1 GDKW01000555 JAI56040.1 ACPB03003844 GAHY01000697 JAA76813.1 GBBI01001521 JAC17191.1 GECZ01003499 JAS66270.1 GL452008 EFN78067.1 GDIP01252196 GDIP01239803 JAI71205.1 GDIP01138956 JAL64758.1 KQ762196 OAD56130.1 GL732551 EFX79490.1 GDIP01132460 LRGB01000944 JAL71254.1 KZS14456.1 GDIP01138955 JAL64759.1 GDIP01151949 JAJ71453.1 GL771866 EFZ09171.1 KK852667 KDR18927.1 GDIQ01097406 JAL54320.1 GDIP01201846 GDIP01137703 JAJ21556.1 BT081534 KV966793 ACO51665.1 PIO23205.1 NEVH01009768 PNF32809.1 GDIQ01015755 JAN78982.1 PYGN01001855 PYGN01000927 PSN32513.1 PSN39303.1 KL206936 KFV87319.1 KB630646 KB631725 KB632417 ERL83764.1 ERL85606.1 ERL95288.1 MUZQ01000333 OWK52629.1 ABQF01093741 GDIP01208997 JAJ14405.1 AHZZ02010894 AQIB01113994 AQIB01113995 APGK01049655 KB741156 ENN73588.1 CM001292 EHH58459.1 JSUE03017739 JU323153 JU473625 CM001269 AFE66909.1 AFH30429.1 EHH28869.1 KQ434943 KZC12204.1 AQIA01029279 AQIA01029280 AQIA01029281 JV667218 AFJ71881.1 ABQF01007637 AAQR03129535 AAQR03129536 AAQR03129537 AAQR03129538 AAQR03129539 KZ288322 PBC28129.1 GL888120 EGI66974.1 GDIP01143258 JAJ80144.1 AK005312 AK010911 AK012083 BC051925 KK718503 KFO57061.1 BC171256 AAI71256.1 AAMC01075323 BC155986 BC156004 AAI55987.1 AAI56005.1 KL504162 KFO84240.1 AC154675 CH466535 EDL36169.1 AGTO01010777 GFXV01000348 MBW12153.1 LZPO01107954 OBS59810.1 KL885896 KGL73381.1 CM004469 OCT93172.1 AAKN02010342 KL216514 KFV69997.1 KL379161 KFP86079.1 AJFE02118924 AJFE02118925 AJFE02118926 AACZ04071644 GABE01010998 NBAG03000041 JAA33741.1 PNI94714.1 KL226060 KFM07594.1 KQ976719 KYM76735.1 AK341148 BAH71605.1 KB321101 ELW47574.1 KK620207 KFV50776.1 KL447391 KFO72876.1 AABR07018059 BC168660 AAI68660.1 KK670381 KFQ04510.1 CABD030087998 BC073617 ADTU01024964 AGTP01099107 AGTP01099108 AGTP01099109 AGTP01099110 AGTP01099111 GABF01005179 GABD01007025 JAA16966.1 JAA26075.1
PZC71850.1 AK403471 BAM19727.1 KQ460205 KPJ17016.1 AGBW02011278 OWR46905.1 GAIX01006022 JAA86538.1 RSAL01000216 RVE44118.1 KF294266 AHZ08394.1 KQ971327 KYB28399.1 NNAY01000195 OXU30096.1 LBMM01001488 KMQ96223.1 GL444789 EFN60586.1 KQ414954 KOC59076.1 GECL01000759 JAP05365.1 GEBQ01027320 JAT12657.1 GDKW01000555 JAI56040.1 ACPB03003844 GAHY01000697 JAA76813.1 GBBI01001521 JAC17191.1 GECZ01003499 JAS66270.1 GL452008 EFN78067.1 GDIP01252196 GDIP01239803 JAI71205.1 GDIP01138956 JAL64758.1 KQ762196 OAD56130.1 GL732551 EFX79490.1 GDIP01132460 LRGB01000944 JAL71254.1 KZS14456.1 GDIP01138955 JAL64759.1 GDIP01151949 JAJ71453.1 GL771866 EFZ09171.1 KK852667 KDR18927.1 GDIQ01097406 JAL54320.1 GDIP01201846 GDIP01137703 JAJ21556.1 BT081534 KV966793 ACO51665.1 PIO23205.1 NEVH01009768 PNF32809.1 GDIQ01015755 JAN78982.1 PYGN01001855 PYGN01000927 PSN32513.1 PSN39303.1 KL206936 KFV87319.1 KB630646 KB631725 KB632417 ERL83764.1 ERL85606.1 ERL95288.1 MUZQ01000333 OWK52629.1 ABQF01093741 GDIP01208997 JAJ14405.1 AHZZ02010894 AQIB01113994 AQIB01113995 APGK01049655 KB741156 ENN73588.1 CM001292 EHH58459.1 JSUE03017739 JU323153 JU473625 CM001269 AFE66909.1 AFH30429.1 EHH28869.1 KQ434943 KZC12204.1 AQIA01029279 AQIA01029280 AQIA01029281 JV667218 AFJ71881.1 ABQF01007637 AAQR03129535 AAQR03129536 AAQR03129537 AAQR03129538 AAQR03129539 KZ288322 PBC28129.1 GL888120 EGI66974.1 GDIP01143258 JAJ80144.1 AK005312 AK010911 AK012083 BC051925 KK718503 KFO57061.1 BC171256 AAI71256.1 AAMC01075323 BC155986 BC156004 AAI55987.1 AAI56005.1 KL504162 KFO84240.1 AC154675 CH466535 EDL36169.1 AGTO01010777 GFXV01000348 MBW12153.1 LZPO01107954 OBS59810.1 KL885896 KGL73381.1 CM004469 OCT93172.1 AAKN02010342 KL216514 KFV69997.1 KL379161 KFP86079.1 AJFE02118924 AJFE02118925 AJFE02118926 AACZ04071644 GABE01010998 NBAG03000041 JAA33741.1 PNI94714.1 KL226060 KFM07594.1 KQ976719 KYM76735.1 AK341148 BAH71605.1 KB321101 ELW47574.1 KK620207 KFV50776.1 KL447391 KFO72876.1 AABR07018059 BC168660 AAI68660.1 KK670381 KFQ04510.1 CABD030087998 BC073617 ADTU01024964 AGTP01099107 AGTP01099108 AGTP01099109 AGTP01099110 AGTP01099111 GABF01005179 GABD01007025 JAA16966.1 JAA26075.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000007151
UP000283053
UP000007266
+ More
UP000215335 UP000036403 UP000000311 UP000053825 UP000192223 UP000002358 UP000015103 UP000008237 UP000000305 UP000076858 UP000027135 UP000235965 UP000245037 UP000053584 UP000001646 UP000030742 UP000197619 UP000007879 UP000007754 UP000028761 UP000029965 UP000233200 UP000233060 UP000019118 UP000009130 UP000006718 UP000076502 UP000233120 UP000233100 UP000233140 UP000233080 UP000005225 UP000242457 UP000007755 UP000000589 UP000052976 UP000008143 UP000233180 UP000016665 UP000233160 UP000092124 UP000005203 UP000189705 UP000189704 UP000053641 UP000186698 UP000005447 UP000053875 UP000240080 UP000002277 UP000053286 UP000002279 UP000078540 UP000007646 UP000011518 UP000053760 UP000002494 UP000001519 UP000005205 UP000005215
UP000215335 UP000036403 UP000000311 UP000053825 UP000192223 UP000002358 UP000015103 UP000008237 UP000000305 UP000076858 UP000027135 UP000235965 UP000245037 UP000053584 UP000001646 UP000030742 UP000197619 UP000007879 UP000007754 UP000028761 UP000029965 UP000233200 UP000233060 UP000019118 UP000009130 UP000006718 UP000076502 UP000233120 UP000233100 UP000233140 UP000233080 UP000005225 UP000242457 UP000007755 UP000000589 UP000052976 UP000008143 UP000233180 UP000016665 UP000233160 UP000092124 UP000005203 UP000189705 UP000189704 UP000053641 UP000186698 UP000005447 UP000053875 UP000240080 UP000002277 UP000053286 UP000002279 UP000078540 UP000007646 UP000011518 UP000053760 UP000002494 UP000001519 UP000005205 UP000005215
Pfam
PF10237 N6-adenineMlase
Interpro
SUPFAM
SSF53335
SSF53335
ProteinModelPortal
H9IU28
A0A2H1VX90
A0A2A4JLY0
A0A2W1BGC8
I4DP87
A0A194RM05
+ More
A0A212EZH5 S4PB98 A0A3S2NN80 A0A067ZWL4 D7EJW9 A0A232FH62 A0A0J7L0V8 E2B162 A0A0L7QKN8 A0A1W4WAJ5 K7J0S4 A0A0V0GD98 A0A1B6KMJ6 A0A0N7Z9H2 R4G8M8 A0A023F731 A0A1B6GV00 E2C1V5 A0A0P4WSG7 A0A0P5SJM6 A0A310SAI0 E9GLC9 A0A0P5T5A5 A0A0P5SJS5 A0A0P5E3W9 E9JD83 A0A067RI41 A0A0P5T1M3 A0A0P5AAG4 C1C3V5 A0A2J7QW40 A0A0P6JKW8 A0A2P8Y4X4 A0A093KAE6 G1KAT8 U4URX5 A0A218UFW3 A0A1X7UYL4 H1A020 A0A0P5A052 A0A096MQR7 A0A0D9RZV1 A0A2K6NLP9 A0A2K5KVP5 N6T7F1 G7PVT6 G7NJQ2 A0A154PL59 A0A2K6DTB8 A0A2K5VXZ2 I2CY46 H0ZM64 A0A2K5XE51 A0A2K5I4G4 H0XTS6 A0A2A3EA95 F4WFJ7 A0A0P5ECT1 Q9CY45 A0A091EJ22 B7ZUJ8 A9JSB0 A0A091GPC2 A0A2K6MVW9 A0A0D2X7Z2 U3K265 A0A2H8TDH0 A0A2K6EJ59 A0A1A6G1X7 A0A087ZRC4 A0A3Q0G2W6 A0A1U7ULI8 A0A099YVG5 A0A1L8HAR5 H0VJ85 A0A093GSI4 A0A091NB18 A0A2R9C8B4 H2Q794 A0A087R290 F7EL08 A0A151HYN4 G3TAT6 C4WUN5 L9JBD9 A0A093F974 A0A091FV71 B5DEG5 A0A091PA60 G3RL82 Q6GN98 A0A158NSJ1 I3MFY8 K7AG74
A0A212EZH5 S4PB98 A0A3S2NN80 A0A067ZWL4 D7EJW9 A0A232FH62 A0A0J7L0V8 E2B162 A0A0L7QKN8 A0A1W4WAJ5 K7J0S4 A0A0V0GD98 A0A1B6KMJ6 A0A0N7Z9H2 R4G8M8 A0A023F731 A0A1B6GV00 E2C1V5 A0A0P4WSG7 A0A0P5SJM6 A0A310SAI0 E9GLC9 A0A0P5T5A5 A0A0P5SJS5 A0A0P5E3W9 E9JD83 A0A067RI41 A0A0P5T1M3 A0A0P5AAG4 C1C3V5 A0A2J7QW40 A0A0P6JKW8 A0A2P8Y4X4 A0A093KAE6 G1KAT8 U4URX5 A0A218UFW3 A0A1X7UYL4 H1A020 A0A0P5A052 A0A096MQR7 A0A0D9RZV1 A0A2K6NLP9 A0A2K5KVP5 N6T7F1 G7PVT6 G7NJQ2 A0A154PL59 A0A2K6DTB8 A0A2K5VXZ2 I2CY46 H0ZM64 A0A2K5XE51 A0A2K5I4G4 H0XTS6 A0A2A3EA95 F4WFJ7 A0A0P5ECT1 Q9CY45 A0A091EJ22 B7ZUJ8 A9JSB0 A0A091GPC2 A0A2K6MVW9 A0A0D2X7Z2 U3K265 A0A2H8TDH0 A0A2K6EJ59 A0A1A6G1X7 A0A087ZRC4 A0A3Q0G2W6 A0A1U7ULI8 A0A099YVG5 A0A1L8HAR5 H0VJ85 A0A093GSI4 A0A091NB18 A0A2R9C8B4 H2Q794 A0A087R290 F7EL08 A0A151HYN4 G3TAT6 C4WUN5 L9JBD9 A0A093F974 A0A091FV71 B5DEG5 A0A091PA60 G3RL82 Q6GN98 A0A158NSJ1 I3MFY8 K7AG74
Ontologies
GO
PANTHER
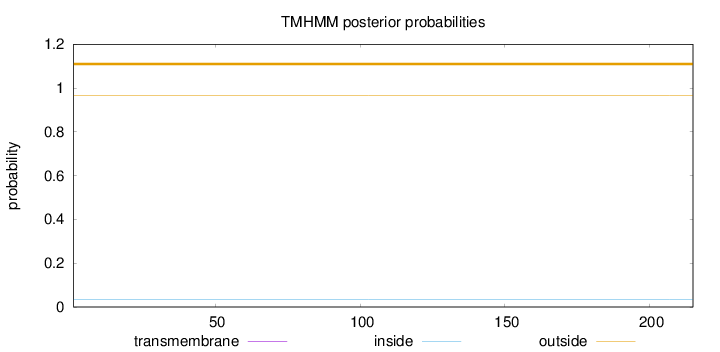
Topology
Subcellular location
Cytoplasm
Length:
215
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00072
Exp number, first 60 AAs:
0.00017
Total prob of N-in:
0.03283
outside
1 - 215
Population Genetic Test Statistics
Pi
295.325942
Theta
201.54042
Tajima's D
1.461944
CLR
11.007557
CSRT
0.784260786960652
Interpretation
Uncertain
