Gene
KWMTBOMO14960
Pre Gene Modal
BGIBMGA000816
Annotation
PREDICTED:_kinesin-like_protein_KIF16B_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 3.298
Sequence
CDS
ATGCCCAGCGAGACCGTCTCCAAGGTCCACCTCGTCGACTTGGCTGGCAGCGAACGTGCTGACGCCACAGGAGCCACAGGGCAGCGGCTGGTAGAAGGCGCTCACATCAATAAGTCGCTGGTCACACTGGGATCCGTCATATCGGCTTTGGCAGAATCCAGTCAGACGGTGGAAATTAAACTACAGAAAGGCTCGAAGAAGAACGTTTTCATTCCGTACCGGGACTCCGTGCTCACGTGGCTCCTGAAGGACTCGCTCGGCGGAAACTCGAAAACTATCATGATAGCCGCGATATCACCGGCGGACTGCAACTACGGCGAAACCCTGTCGACCCTGCGGTACGCGAACCGCGCCAAGAACATCATCAACAAGCCGACCATCAACGAGGATCCCAACGTCAAGCTCATCCGCGAGCTGCGCGAGGAGATCGACAAGCTTCGGGCGCAGCTCACGCACAATTCGTGCTCTGTTGAGAACGAGCCGGGAGTGCTGGCGACGTTGCAGCGCAAAGAAGCCCAAGAGAAGGTCCTCACAGAGAAGTGGACGGAGAAGTGGCGAGAAACGCAACAGATCCTTCAGGAACAGAAAGCTCTCGGACTCAGGAAGAGCGGACTCGGCGTGGTGCTGGACTCGGACATGCCGCACCTCGTGGGGATCGACGACAACCTGCTGTCCACCGGCGTCACGCTCTACCACCTCAAGGAAGGCGAGACGCTGATAGGCACGGAGGAGCACTCGCCGATACCGGACATCGTGCTGTCGGGCCCCGGCGTGCTGCCGCTGCACTGCCGCGTGACGCTGTGCGCCGGCACCGCCACGCTGCACCCCTGTGCCGGCGCGCACTGCTGGCTCAACACCGTGCTGCTCGACAAGCCGGCCAAACTCAGCCAAGGTTGCATTCTTCTCCTGGGCCGAACCAACATGTTCCGGTACAACGACCCCGCGGAGGCGGCCAAGCTGCGCCAGGAGGGCAGCGCCAACATGAACCTGTCTCGGCTCTCCTTGCTGTCCTGGTCCACCACCGACCTCGCGGCCAGCACCGAGAACCTCAACTCCAGCGCCCTCTCGGATCTAGAAAGCGAGATCCGTTGCGAGCGTATAGAGGCACAGCGCGCCGAGCTGGAGCGAGAGAGACAACAGTTCCTGATGCAGCAGGAGGAGCGCCAGCGCAGGTGGAACGCGGAGAGGGAGGAGCTCCTGCAGGCGCAAAAGAAACTTGACGAGGAACGCGAGGCGATGGAGAAGGAATACGCGGCGGCCTGCAGGAGACTGTCCGGGGACTGGAGGGCGCTGGAGCGGGGCTGGACCAACCGCAGGAGGGCGCTGCGCTGCAGGCAGAGGGAGCTGGCGGCCAGGACGCAGAGACTCAGGGGGCTCAGCGACACTCATCAGGGATGTGTGGAGACCGAGGAATCGCGCATGCTGGAACTCAGATCGAGGATCGAAAACAAGAGGATAGAGTTGGAGGACTACGCCAAGAATATCATCAGGGAATTAGTGGAGAGCGGCAAAATCGAGAGTTCAACGCCAAGTCCGGACAGTCCCGCATCGGAGACCAGCACAGAGAGCCTGATGCACCTGCTGCGTCAGTGCGAGCCCGACACCGCGACCAGCATCAAGGACACCGTCGACCGGCATAAGAGAGAGCTGGTGGAGCTGGAGGAGGAGCTGCAGAGCCGGGTGCGGTCGGTGTCGAACCAGCGCGTGAAGGTGGAACGGCTGCAGGCCGAGCTGGCGCTGCTGGCCGTGCAGGAGCTGGGCCTGCGCCGCGCGCTCGCCGCCGACCTCACGCCCGCCGCGCCCACAGAAGGTGACAGTCAAGCGTCGCCCACGGAAGGAGTGAAGCGCAGCAAGTCCGAGCTGGTGCTGCGCAGGACTCCTCGCGAGGAGGTGCTCGACCCACTCTCCGCTGATGAACGAGATGAATTCCGGGACAAAAGAATCAACCTCAGTCTGAGTCTAGATCCTCTGGCGTGTCCCGACTCGCTGAACGCGGAGGAGTACCAGACCGCGAGCTCCACCACCCCGAAGTCATTCCCCCCGGAGAGCCCGCCCCCCCGCAGCTCCCCGCAGAGCTCGCCCCCCCGCGGCCCCCCGCAGCCGAACTTCCGGCTGGGCCCCGTGCCCGACGACAGCGACGATATGTCCAGCACTGAAGACTATCACAAGCCCAGGATAGTGGAGGGCCAGTCGTCAAGCTCCATGGACAGGTCGCCGGAAGAGAGACACCAGAGAGCAAAATACAGAAGAAAGATCACTGGCGACAGTAAGTCGGGGCGCCGACAAGTGACCGAGGCAGAAGCACTGCGCGCGATGCAGAGGCTGTGCTCTCGGATAGCTTCACAGAAGATGCTCGTCATCTCTTCACTGGAGAACGACTGCTCGAAGGAGGAACTAAACAGGCAGATTGCGGTTCTCCAAGAGCTTCAAAAGAAATACGTCAGGCTGGAGATGGCGCTGCAGTATTCGTTCTTCGACAGCCCGCAGAACAGGAGGTCGATGATGGTCACTCCGATACAGGAAACTCCCTCCCTCACCCTCTCCGAGAGCGACATGCAGGTCGCTACGGACGACCCCGGCGCCGAGAACGGGAACGGAGTCCTCACCGACCCTCTGCTCAACAGGCTGAAGGAGCAGGAGCTGGAGGCGTCGGACAACGTGTCGACGTCCAACGACGAGCTGCCCGCGGAGCAGCCGGACGCGTGGGACGACCCGCTGCGCCGCGCGCTGGAGCCCGACGCGCCCGCACGCAACGGAGCCGTCGCGCCCACGCTCGTGCATCTACCGCTCGAGGAATCAACAGACAACGGAGGTCTGACTCACCCCATAGATTTTAACGCGGTGGTGAGCGTGCCGGGGTGGGTGACGCGCGGGGCGGGGGCGCGCACGCACCACGAGTACGAGGTGCGCGTGTGGCTGGGCGCGCGCTGTCGGTACTCGCTGCTGCGGCGCTACCGGCGCTTCCGGGACCTCTACCTCGAGATGCGGGCGCGCTACCCGGCGCAGGTGCGTGCTACTCACACACAGCCAATGAAACAACGTTCTAATCTGATTTCCCTCTATGTTTAG
Protein
MPSETVSKVHLVDLAGSERADATGATGQRLVEGAHINKSLVTLGSVISALAESSQTVEIKLQKGSKKNVFIPYRDSVLTWLLKDSLGGNSKTIMIAAISPADCNYGETLSTLRYANRAKNIINKPTINEDPNVKLIRELREEIDKLRAQLTHNSCSVENEPGVLATLQRKEAQEKVLTEKWTEKWRETQQILQEQKALGLRKSGLGVVLDSDMPHLVGIDDNLLSTGVTLYHLKEGETLIGTEEHSPIPDIVLSGPGVLPLHCRVTLCAGTATLHPCAGAHCWLNTVLLDKPAKLSQGCILLLGRTNMFRYNDPAEAAKLRQEGSANMNLSRLSLLSWSTTDLAASTENLNSSALSDLESEIRCERIEAQRAELERERQQFLMQQEERQRRWNAEREELLQAQKKLDEEREAMEKEYAAACRRLSGDWRALERGWTNRRRALRCRQRELAARTQRLRGLSDTHQGCVETEESRMLELRSRIENKRIELEDYAKNIIRELVESGKIESSTPSPDSPASETSTESLMHLLRQCEPDTATSIKDTVDRHKRELVELEEELQSRVRSVSNQRVKVERLQAELALLAVQELGLRRALAADLTPAAPTEGDSQASPTEGVKRSKSELVLRRTPREEVLDPLSADERDEFRDKRINLSLSLDPLACPDSLNAEEYQTASSTTPKSFPPESPPPRSSPQSSPPRGPPQPNFRLGPVPDDSDDMSSTEDYHKPRIVEGQSSSSMDRSPEERHQRAKYRRKITGDSKSGRRQVTEAEALRAMQRLCSRIASQKMLVISSLENDCSKEELNRQIAVLQELQKKYVRLEMALQYSFFDSPQNRRSMMVTPIQETPSLTLSESDMQVATDDPGAENGNGVLTDPLLNRLKEQELEASDNVSTSNDELPAEQPDAWDDPLRRALEPDAPARNGAVAPTLVHLPLEESTDNGGLTHPIDFNAVVSVPGWVTRGAGARTHHEYEVRVWLGARCRYSLLRRYRRFRDLYLEMRARYPAQVRATHTQPMKQRSNLISLYV
Summary
Similarity
Belongs to the TRAFAC class myosin-kinesin ATPase superfamily. Kinesin family.
Uniprot
EMBL
Proteomes
Interpro
Gene 3D
CDD
ProteinModelPortal
PDB
6A20
E-value=2.24398e-49,
Score=498
Ontologies
GO
PANTHER
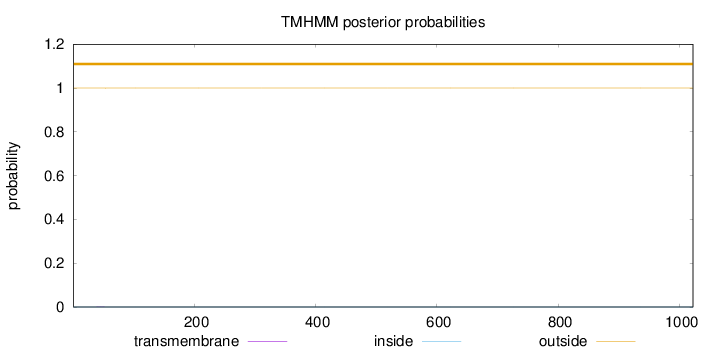
Topology
Length:
1022
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00495999999999998
Exp number, first 60 AAs:
0.00338
Total prob of N-in:
0.00022
outside
1 - 1022
Population Genetic Test Statistics
Pi
265.451762
Theta
168.144238
Tajima's D
0.825292
CLR
0.529508
CSRT
0.604469776511174
Interpretation
Uncertain
