Gene
KWMTBOMO14959
Pre Gene Modal
BGIBMGA000816
Annotation
PREDICTED:_kinesin-like_protein_KIF16B_[Papilio_xuthus]
Full name
Kinesin-like protein
+ More
Kinesin-like protein Klp98A
Kinesin-like protein Klp98A
Location in the cell
Cytoplasmic Reliability : 1.347 Mitochondrial Reliability : 1.083 Nuclear Reliability : 1.294
Sequence
CDS
ATGACAATGGCCTCGGTAAAAGTTGCTGTAAGGGTTCGCCCGTTCAATCAAAGAGAAAAGGACATGGCCGCCAAACTCATAGTTCAGATGGAGGGAAAGAAAACGCGACTCCTTACCGTCAAAAGTAGCAAGGAACAAAATGAGGGTAGCCGTGAAAAGTACAAGGACTTCACGTTCGATCATTCTTACTGGTCATTCGACGGCGGCGACAAGCATTACGCTTCACAAGAACAGGTATTTTCCGACTTGGGTCTGGATGTGATAGACAGCGCTTTCGAAGGATACAATGCCTGTGTTTTCGCTTACGGTCAAACTGGAAGCGGGAAGACTTTCACCATGATGGGATCGCCGGAATGCCAAGGTCTGATACCGCGCATATGCCGGCAACTATTCTCCCGCGTGGCAGCCGGCAAGGAGTCAGGGGCTTCCTACCGCACCGAGGTCAGCTACTTAGAGATCTACAACGAAAGAGTGAAAGATCTGTTGTCAACGGATGCGGGACACTCCCTCAGGGTCAGGGAGCATCCGAAGCTGGGACCGTACGTTCAGGATCTGTCCAAGCACTTGGTCTCCGATTATGATGATATACAGGTGAGCAATTCAATATATGTAAACTGTACAGGAATAAAGAGATGA
Protein
MTMASVKVAVRVRPFNQREKDMAAKLIVQMEGKKTRLLTVKSSKEQNEGSREKYKDFTFDHSYWSFDGGDKHYASQEQVFSDLGLDVIDSAFEGYNACVFAYGQTGSGKTFTMMGSPECQGLIPRICRQLFSRVAAGKESGASYRTEVSYLEIYNERVKDLLSTDAGHSLRVREHPKLGPYVQDLSKHLVSDYDDIQVSNSIYVNCTGIKR
Summary
Description
Plus end-directed motor protein involved in asymmetric cell division of sensory organ precursor (SOP) cells by playing a role in the asymmetric localization of Sara-expressing endosomes to the pIIa daughter cell but not to the pIIb cell. Targets Sara-expressing endosomes to the central spindle which is symmetrically arranged in early cell division. During late cytokinesis, central spindle asymmetry is generated by enrichment of Patronin on the pIIb side which protects microtubules from depolymerization by Klp10A while unprotected microtubules on the pIIa side are disassembled by Klp10A, leading to the asymmetric delivery of Sara-expressing endosomes to the pIIa daughter cell (PubMed:26659188). Also plays a role in regulation of autophagosome formation, fusion and positioning and is required for normal localization of Rab14 (PubMed:26763909).
Subunit
Interacts with Atg8a and Rab14.
Similarity
Belongs to the TRAFAC class myosin-kinesin ATPase superfamily. Kinesin family.
Keywords
Alternative splicing
ATP-binding
Autophagy
Coiled coil
Complete proteome
Endosome
Microtubule
Motor protein
Nucleotide-binding
Reference proteome
Transport
Feature
chain Kinesin-like protein Klp98A
splice variant In isoform B.
splice variant In isoform B.
Uniprot
A0A212EZN6
A0A2A4JGL4
A0A2A4JFR8
A0A194PFN7
A0A2W1BJL2
H9IU86
+ More
A0A2J7PR34 A0A2A3ED30 A0A087ZTB7 A0A2H1VPS9 A0A232EWS6 A0A0C9R9J7 A0A3L8E2Q4 E2BSL5 A0A151JTB6 A0A2A4JFQ6 K7J3W7 A0A195DCZ0 A0A158NMP5 A0A1B6JJ36 A0A0C9QMU2 A0A226ETH9 A0A1B6EFZ0 F4X310 A0A151X1K7 A0A1B6KSV7 A0A0Q9XA93 A0A0Q9XBH7 B4KDX5 A0A0N8P1P8 B3LXJ5 B4NAQ0 B4JTD4 A0A034VYU4 B4G2L1 A0A0M5J1D2 A0A0R3NG26 Q29A93 A0A034W2C1 A0A3B0K3T0 A0A1W4URR2 A0A1W4UDB0 B3P603 Q9VB25-2 A0A0R1EA50 B4QXH5 A0A1W4UPV8 A0A1W4UQ51 Q9VB25 A0A3B0K4T6 B4PRQ1 A0A026WKY9 A0A0K8UNF4 E2AIY1 A0A0K8VF26 A0A0A1WE10 A0A1I8Q073 B4IGT3 A0A151I5Y6 A0A194RHV4 A0A1I8M6T3 A0A131XYU4 A0A1I8M6U7 A0A182KMW9 Q7PML3 B4M030 A0A1E1XEU5 A0A0K8TKQ0 A0A3R7LZJ8 A7SRC2 A0A084W920 A0A0T6AVX8 A0A3M6UGV7 A0A131YA00 A0A224YS54 A0A2B4RU09 A0A1A9WYT0 A0A2R5LPK0 W8AM04 W8AYA5 A0A1E1XLW9 A0A0P4WG05 A0A1J1J9V3 A0A131YJ83 L7M6K4 A0A0N0U647 T1JPN4 D6WQE7 A0A195C7R9 A0A1D2NAT5 S4RH81 A0A1Y1MTK4 R7TZ34 A0A2G8K9K7 A0A1B0FAF6 A0A1B0BUI8 A0A210QWK5 A0A1A9XIU4 A0A1A9ZAW3 A0A1A9US86
A0A2J7PR34 A0A2A3ED30 A0A087ZTB7 A0A2H1VPS9 A0A232EWS6 A0A0C9R9J7 A0A3L8E2Q4 E2BSL5 A0A151JTB6 A0A2A4JFQ6 K7J3W7 A0A195DCZ0 A0A158NMP5 A0A1B6JJ36 A0A0C9QMU2 A0A226ETH9 A0A1B6EFZ0 F4X310 A0A151X1K7 A0A1B6KSV7 A0A0Q9XA93 A0A0Q9XBH7 B4KDX5 A0A0N8P1P8 B3LXJ5 B4NAQ0 B4JTD4 A0A034VYU4 B4G2L1 A0A0M5J1D2 A0A0R3NG26 Q29A93 A0A034W2C1 A0A3B0K3T0 A0A1W4URR2 A0A1W4UDB0 B3P603 Q9VB25-2 A0A0R1EA50 B4QXH5 A0A1W4UPV8 A0A1W4UQ51 Q9VB25 A0A3B0K4T6 B4PRQ1 A0A026WKY9 A0A0K8UNF4 E2AIY1 A0A0K8VF26 A0A0A1WE10 A0A1I8Q073 B4IGT3 A0A151I5Y6 A0A194RHV4 A0A1I8M6T3 A0A131XYU4 A0A1I8M6U7 A0A182KMW9 Q7PML3 B4M030 A0A1E1XEU5 A0A0K8TKQ0 A0A3R7LZJ8 A7SRC2 A0A084W920 A0A0T6AVX8 A0A3M6UGV7 A0A131YA00 A0A224YS54 A0A2B4RU09 A0A1A9WYT0 A0A2R5LPK0 W8AM04 W8AYA5 A0A1E1XLW9 A0A0P4WG05 A0A1J1J9V3 A0A131YJ83 L7M6K4 A0A0N0U647 T1JPN4 D6WQE7 A0A195C7R9 A0A1D2NAT5 S4RH81 A0A1Y1MTK4 R7TZ34 A0A2G8K9K7 A0A1B0FAF6 A0A1B0BUI8 A0A210QWK5 A0A1A9XIU4 A0A1A9ZAW3 A0A1A9US86
Pubmed
22118469
26354079
28756777
19121390
28648823
30249741
+ More
20798317 20075255 21347285 21719571 17994087 25348373 15632085 10731132 12537572 12537569 1924306 26659188 26763909 17550304 24508170 25830018 25315136 20966253 12364791 14747013 17210077 28503490 26369729 17615350 24438588 30382153 26830274 28797301 24495485 29209593 25576852 18362917 19820115 27289101 28004739 23254933 29023486 28812685
20798317 20075255 21347285 21719571 17994087 25348373 15632085 10731132 12537572 12537569 1924306 26659188 26763909 17550304 24508170 25830018 25315136 20966253 12364791 14747013 17210077 28503490 26369729 17615350 24438588 30382153 26830274 28797301 24495485 29209593 25576852 18362917 19820115 27289101 28004739 23254933 29023486 28812685
EMBL
AGBW02011278
OWR46904.1
NWSH01001542
PCG70916.1
PCG70917.1
KQ459604
+ More
KPI92102.1 KZ150248 PZC71853.1 BABH01034241 BABH01034242 NEVH01022633 PNF18789.1 KZ288292 PBC29119.1 ODYU01003717 SOQ42845.1 NNAY01001825 OXU22796.1 GBYB01004830 JAG74597.1 QOIP01000001 RLU27021.1 GL450241 EFN81261.1 KQ981936 KYN32776.1 PCG70915.1 AAZX01006133 AAZX01009857 KQ981010 KYN10304.1 ADTU01020662 ADTU01020663 ADTU01020664 GECU01008502 GECU01005620 JAS99204.1 JAT02087.1 GBYB01004829 JAG74596.1 LNIX01000002 OXA60508.1 GEDC01000450 JAS36848.1 GL888604 EGI59168.1 KQ982588 KYQ54134.1 GEBQ01025633 JAT14344.1 CH933806 KRG01303.1 KRG01304.1 EDW14972.1 CH902617 KPU80524.1 EDV43889.1 CH964232 EDW80864.1 CH916373 EDV95024.1 GAKP01010456 JAC48496.1 CH479179 EDW24056.1 CP012526 ALC45561.1 CM000070 KRT00039.1 EAL27457.1 GAKP01010455 JAC48497.1 OUUW01000005 SPP80639.1 CH954182 EDV53403.1 AE014297 AY070884 BT132947 M74432 AAA28659.1 AAL48506.1 AEW43887.1 CM000160 KRK04306.1 CM000364 EDX14661.1 SPP80636.1 EDW98494.1 KK107159 EZA56695.1 GDHF01024276 JAI28038.1 GL439921 EFN66570.1 GDHF01014833 JAI37481.1 GBXI01017150 GBXI01008102 JAC97141.1 JAD06190.1 CH480837 EDW49051.1 KQ976431 KYM87619.1 KQ460205 KPJ17019.1 GEFM01004368 JAP71428.1 AAAB01008978 EAA13643.5 CH940650 EDW68280.1 GFAC01001480 JAT97708.1 GDAI01002897 JAI14706.1 QCYY01002627 ROT68838.1 DS469760 EDO33740.1 ATLV01021573 KE525320 KFB46714.1 LJIG01022678 KRT79292.1 RCHS01001558 RMX52876.1 GEDV01012468 JAP76089.1 GFPF01009260 MAA20406.1 LSMT01000346 PFX19735.1 GGLE01007111 MBY11237.1 GAMC01016730 GAMC01016728 JAB89825.1 GAMC01016729 GAMC01016727 JAB89828.1 GFAA01003443 JAT99991.1 GDRN01025491 JAI67719.1 CVRI01000073 CRL07801.1 GEDV01010047 JAP78510.1 GACK01005369 JAA59665.1 KQ435736 KOX77052.1 CAEY01000428 KQ971354 EFA06082.2 KQ978231 KYM96211.1 LJIJ01000115 ODN02361.1 GEZM01024261 JAV87820.1 AMQN01010291 KB307500 ELT98872.1 MRZV01000763 PIK44649.1 CCAG010019388 JXJN01020709 JXJN01020710 NEDP02001506 OWF53130.1
KPI92102.1 KZ150248 PZC71853.1 BABH01034241 BABH01034242 NEVH01022633 PNF18789.1 KZ288292 PBC29119.1 ODYU01003717 SOQ42845.1 NNAY01001825 OXU22796.1 GBYB01004830 JAG74597.1 QOIP01000001 RLU27021.1 GL450241 EFN81261.1 KQ981936 KYN32776.1 PCG70915.1 AAZX01006133 AAZX01009857 KQ981010 KYN10304.1 ADTU01020662 ADTU01020663 ADTU01020664 GECU01008502 GECU01005620 JAS99204.1 JAT02087.1 GBYB01004829 JAG74596.1 LNIX01000002 OXA60508.1 GEDC01000450 JAS36848.1 GL888604 EGI59168.1 KQ982588 KYQ54134.1 GEBQ01025633 JAT14344.1 CH933806 KRG01303.1 KRG01304.1 EDW14972.1 CH902617 KPU80524.1 EDV43889.1 CH964232 EDW80864.1 CH916373 EDV95024.1 GAKP01010456 JAC48496.1 CH479179 EDW24056.1 CP012526 ALC45561.1 CM000070 KRT00039.1 EAL27457.1 GAKP01010455 JAC48497.1 OUUW01000005 SPP80639.1 CH954182 EDV53403.1 AE014297 AY070884 BT132947 M74432 AAA28659.1 AAL48506.1 AEW43887.1 CM000160 KRK04306.1 CM000364 EDX14661.1 SPP80636.1 EDW98494.1 KK107159 EZA56695.1 GDHF01024276 JAI28038.1 GL439921 EFN66570.1 GDHF01014833 JAI37481.1 GBXI01017150 GBXI01008102 JAC97141.1 JAD06190.1 CH480837 EDW49051.1 KQ976431 KYM87619.1 KQ460205 KPJ17019.1 GEFM01004368 JAP71428.1 AAAB01008978 EAA13643.5 CH940650 EDW68280.1 GFAC01001480 JAT97708.1 GDAI01002897 JAI14706.1 QCYY01002627 ROT68838.1 DS469760 EDO33740.1 ATLV01021573 KE525320 KFB46714.1 LJIG01022678 KRT79292.1 RCHS01001558 RMX52876.1 GEDV01012468 JAP76089.1 GFPF01009260 MAA20406.1 LSMT01000346 PFX19735.1 GGLE01007111 MBY11237.1 GAMC01016730 GAMC01016728 JAB89825.1 GAMC01016729 GAMC01016727 JAB89828.1 GFAA01003443 JAT99991.1 GDRN01025491 JAI67719.1 CVRI01000073 CRL07801.1 GEDV01010047 JAP78510.1 GACK01005369 JAA59665.1 KQ435736 KOX77052.1 CAEY01000428 KQ971354 EFA06082.2 KQ978231 KYM96211.1 LJIJ01000115 ODN02361.1 GEZM01024261 JAV87820.1 AMQN01010291 KB307500 ELT98872.1 MRZV01000763 PIK44649.1 CCAG010019388 JXJN01020709 JXJN01020710 NEDP02001506 OWF53130.1
Proteomes
UP000007151
UP000218220
UP000053268
UP000005204
UP000235965
UP000242457
+ More
UP000005203 UP000215335 UP000279307 UP000008237 UP000078541 UP000002358 UP000078492 UP000005205 UP000198287 UP000007755 UP000075809 UP000009192 UP000007801 UP000007798 UP000001070 UP000008744 UP000092553 UP000001819 UP000268350 UP000192221 UP000008711 UP000000803 UP000002282 UP000000304 UP000053097 UP000000311 UP000095300 UP000001292 UP000078540 UP000053240 UP000095301 UP000075882 UP000007062 UP000008792 UP000283509 UP000001593 UP000030765 UP000275408 UP000225706 UP000091820 UP000183832 UP000053105 UP000015104 UP000007266 UP000078542 UP000094527 UP000245300 UP000014760 UP000230750 UP000092444 UP000092460 UP000242188 UP000092443 UP000092445 UP000078200
UP000005203 UP000215335 UP000279307 UP000008237 UP000078541 UP000002358 UP000078492 UP000005205 UP000198287 UP000007755 UP000075809 UP000009192 UP000007801 UP000007798 UP000001070 UP000008744 UP000092553 UP000001819 UP000268350 UP000192221 UP000008711 UP000000803 UP000002282 UP000000304 UP000053097 UP000000311 UP000095300 UP000001292 UP000078540 UP000053240 UP000095301 UP000075882 UP000007062 UP000008792 UP000283509 UP000001593 UP000030765 UP000275408 UP000225706 UP000091820 UP000183832 UP000053105 UP000015104 UP000007266 UP000078542 UP000094527 UP000245300 UP000014760 UP000230750 UP000092444 UP000092460 UP000242188 UP000092443 UP000092445 UP000078200
Interpro
Gene 3D
CDD
ProteinModelPortal
A0A212EZN6
A0A2A4JGL4
A0A2A4JFR8
A0A194PFN7
A0A2W1BJL2
H9IU86
+ More
A0A2J7PR34 A0A2A3ED30 A0A087ZTB7 A0A2H1VPS9 A0A232EWS6 A0A0C9R9J7 A0A3L8E2Q4 E2BSL5 A0A151JTB6 A0A2A4JFQ6 K7J3W7 A0A195DCZ0 A0A158NMP5 A0A1B6JJ36 A0A0C9QMU2 A0A226ETH9 A0A1B6EFZ0 F4X310 A0A151X1K7 A0A1B6KSV7 A0A0Q9XA93 A0A0Q9XBH7 B4KDX5 A0A0N8P1P8 B3LXJ5 B4NAQ0 B4JTD4 A0A034VYU4 B4G2L1 A0A0M5J1D2 A0A0R3NG26 Q29A93 A0A034W2C1 A0A3B0K3T0 A0A1W4URR2 A0A1W4UDB0 B3P603 Q9VB25-2 A0A0R1EA50 B4QXH5 A0A1W4UPV8 A0A1W4UQ51 Q9VB25 A0A3B0K4T6 B4PRQ1 A0A026WKY9 A0A0K8UNF4 E2AIY1 A0A0K8VF26 A0A0A1WE10 A0A1I8Q073 B4IGT3 A0A151I5Y6 A0A194RHV4 A0A1I8M6T3 A0A131XYU4 A0A1I8M6U7 A0A182KMW9 Q7PML3 B4M030 A0A1E1XEU5 A0A0K8TKQ0 A0A3R7LZJ8 A7SRC2 A0A084W920 A0A0T6AVX8 A0A3M6UGV7 A0A131YA00 A0A224YS54 A0A2B4RU09 A0A1A9WYT0 A0A2R5LPK0 W8AM04 W8AYA5 A0A1E1XLW9 A0A0P4WG05 A0A1J1J9V3 A0A131YJ83 L7M6K4 A0A0N0U647 T1JPN4 D6WQE7 A0A195C7R9 A0A1D2NAT5 S4RH81 A0A1Y1MTK4 R7TZ34 A0A2G8K9K7 A0A1B0FAF6 A0A1B0BUI8 A0A210QWK5 A0A1A9XIU4 A0A1A9ZAW3 A0A1A9US86
A0A2J7PR34 A0A2A3ED30 A0A087ZTB7 A0A2H1VPS9 A0A232EWS6 A0A0C9R9J7 A0A3L8E2Q4 E2BSL5 A0A151JTB6 A0A2A4JFQ6 K7J3W7 A0A195DCZ0 A0A158NMP5 A0A1B6JJ36 A0A0C9QMU2 A0A226ETH9 A0A1B6EFZ0 F4X310 A0A151X1K7 A0A1B6KSV7 A0A0Q9XA93 A0A0Q9XBH7 B4KDX5 A0A0N8P1P8 B3LXJ5 B4NAQ0 B4JTD4 A0A034VYU4 B4G2L1 A0A0M5J1D2 A0A0R3NG26 Q29A93 A0A034W2C1 A0A3B0K3T0 A0A1W4URR2 A0A1W4UDB0 B3P603 Q9VB25-2 A0A0R1EA50 B4QXH5 A0A1W4UPV8 A0A1W4UQ51 Q9VB25 A0A3B0K4T6 B4PRQ1 A0A026WKY9 A0A0K8UNF4 E2AIY1 A0A0K8VF26 A0A0A1WE10 A0A1I8Q073 B4IGT3 A0A151I5Y6 A0A194RHV4 A0A1I8M6T3 A0A131XYU4 A0A1I8M6U7 A0A182KMW9 Q7PML3 B4M030 A0A1E1XEU5 A0A0K8TKQ0 A0A3R7LZJ8 A7SRC2 A0A084W920 A0A0T6AVX8 A0A3M6UGV7 A0A131YA00 A0A224YS54 A0A2B4RU09 A0A1A9WYT0 A0A2R5LPK0 W8AM04 W8AYA5 A0A1E1XLW9 A0A0P4WG05 A0A1J1J9V3 A0A131YJ83 L7M6K4 A0A0N0U647 T1JPN4 D6WQE7 A0A195C7R9 A0A1D2NAT5 S4RH81 A0A1Y1MTK4 R7TZ34 A0A2G8K9K7 A0A1B0FAF6 A0A1B0BUI8 A0A210QWK5 A0A1A9XIU4 A0A1A9ZAW3 A0A1A9US86
PDB
2HXH
E-value=3.52353e-53,
Score=523
Ontologies
GO
PANTHER
Topology
Subcellular location
Early endosome
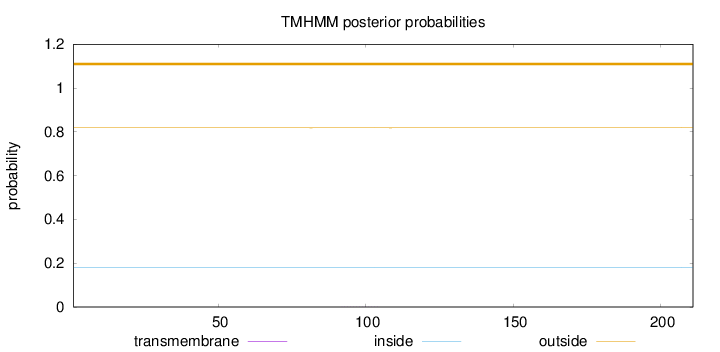
Length:
211
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00315
Exp number, first 60 AAs:
0
Total prob of N-in:
0.18169
outside
1 - 211
Population Genetic Test Statistics
Pi
24.050564
Theta
26.80106
Tajima's D
-1.130271
CLR
0.733684
CSRT
0.112744362781861
Interpretation
Uncertain
