Gene
KWMTBOMO14956
Pre Gene Modal
BGIBMGA000814
Annotation
PREDICTED:_bromodomain_adjacent_to_zinc_finger_domain_protein_1A_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.288
Sequence
CDS
ATGGACACGAACTCGCGCGCCTACTGTACATTTTCACTTCATTTGCTGCACCTGCTGTCGTCGGGCGCGGCGGGCACGGCGCGGGGCGCGGCGTGGCGCGCGTCGCAGCGCGGCGGCTACTCCTCGCTCGACGACCCCGGCCTGCGACTGCGCCGGCGCGCGCCACAGCTGCTCGTGCGGCTCGCCAAGTACCACGTGGCTGAGCTCGGACTAGATGACAAATTCGAAATACTCCAGTGCTTGATGAACCAGATACTGACTTACGCGACCGTCCGTGATGTCATTGAGGAAAAACTCGAGGAACACAAGAATTCTAAGCAAGCTTTGAGACTTTTACAGATAAACGAAAGGAAACGCGAAGCGCAACTGCTCGCGTCGAAACAAGATCTTAAGAAAGAAGCTGCAGCTAAAAAAGAGGAGAACAAGCTCTCTGGTGAAAAAGCTCGAGCTATAGACGAACAGCTTAAAGTCGACACAGAGAAGCTGAACAAAGAAAACGACGAGAAAAAGAAAGAGTACGAAAAGAAATTAAAAGAACTGCAAATCGGATTGTTCGACTATTCTTCTTATCTTGGCATGGATCGCGCTTACCGTCGCTACTGGCTCAACCAGGCCGTGGCCGGGCTGTTCGTGGAGGCGGGCACGGAGCCGCGCGGGCCGTGCCGCGACAAGCCGCTGCCTTCCGCTCCTGAGCACGGAGAGGACACCCTCACCTATGTCACTAGATTGTTTGAAACCGAAAAGGAAAGAGCGAGTAGTGATAAAGAAAATGATTCGGCAGCAAATTCACGGGGGAATTCCCCAAAGAAACCGCTCACGAACATCAATGGCTTAACACACAGGAACGGGTTCGATGATATCACACAACAGCTATTGATCTGCAGCGGAGATCTTTCTACGTGCAAAGTGCATGGAAAGGTCGACCGGCCCCAATGGTGGGTCTACCACACAGAGGAACAGATAGAAGCATTGATCCAGTCTCTAAACAAGCGCGGTATTAGGGAAAGTGAACTCCGACAGAGCCTGGAGCTGGACAAGGACAACATCATTCAGTACCTGAGGAAATGTCCTGTCAAGTACCTCAATGGATCTGCTGCTAGTTCTCCCCCTCCCCCCGCGCCGCACTACACGCGCCGCCGCCCCCCGCCCCCCTCGCTCACCGTGCCGCGCGACTCCTCGCTGGCCGAGGCCGTGGAGCTGGTACTGCGCGACTATATCCTCGATCTGGAGGAGAAGATTCACTACGGATGCCTTGGAGCCCTTAAAGTTAAGGACCGCGAGGCGTGGCGCGGGACGATAATGCTGCGCGGGTACGACAAACAAGCCGACTACCTGACCTGGGGCCCCAACCAAATGTACAGAGATGACTATCACCAGCCCAACGGCGTTCTGAATATTCCTCAAGATCTTGACGAAACAGAGCTAGAGTCGATACCGGTGAACAAATACCGCGACCCCGGCTACTACCTGGAGGCCGCCAGGGTGAACGGAGTCAAGGTAGAGGGCGACGAGCTGAAGGCGAGGCGTGACGTCATCCGAGGGCTCGCCTGCGCCCTGCTGCAGGTCTCCCAGGCCATACACGCCAAATACTTGAAACGGCCTCTGGGGTGGGACGAGAAGGGGCGGGCGCGCAGCACGGAGGGTGGCGCGCTGGCCCGCTGGCAGGTGTCGCTGCTGGAGTGCGGCAGCGCGGCGGGCGCGCGGCTGCACGCGGCGGCGCTGCACGCGTCCGTGTGCTGGCGCGCGTCCGTGCTGCGCGCCGCCTGCCGCCTGTGCCGCCGCCGCGCCGACCCGCACGCCATGCTGCTGTGCGACGCCTGCAACGCCGGCCACCACCTCTACTGCCTCACGCCGCCGCTGCAGGTACAAGCAGTTGAGAAGTGGCAAGGGACACCCGGATGGAACGAAACTCCTTTCGATTAA
Protein
MDTNSRAYCTFSLHLLHLLSSGAAGTARGAAWRASQRGGYSSLDDPGLRLRRRAPQLLVRLAKYHVAELGLDDKFEILQCLMNQILTYATVRDVIEEKLEEHKNSKQALRLLQINERKREAQLLASKQDLKKEAAAKKEENKLSGEKARAIDEQLKVDTEKLNKENDEKKKEYEKKLKELQIGLFDYSSYLGMDRAYRRYWLNQAVAGLFVEAGTEPRGPCRDKPLPSAPEHGEDTLTYVTRLFETEKERASSDKENDSAANSRGNSPKKPLTNINGLTHRNGFDDITQQLLICSGDLSTCKVHGKVDRPQWWVYHTEEQIEALIQSLNKRGIRESELRQSLELDKDNIIQYLRKCPVKYLNGSAASSPPPPAPHYTRRRPPPPSLTVPRDSSLAEAVELVLRDYILDLEEKIHYGCLGALKVKDREAWRGTIMLRGYDKQADYLTWGPNQMYRDDYHQPNGVLNIPQDLDETELESIPVNKYRDPGYYLEAARVNGVKVEGDELKARRDVIRGLACALLQVSQAIHAKYLKRPLGWDEKGRARSTEGGALARWQVSLLECGSAAGARLHAAALHASVCWRASVLRAACRLCRRRADPHAMLLCDACNAGHHLYCLTPPLQVQAVEKWQGTPGWNETPFD
Summary
Uniprot
H9IU84
A0A2A4JG07
A0A2H1VPS5
A0A212EZJ2
A0A194PHB9
A0A194RI66
+ More
A0A2J7R258 A0A2J7R251 A0A2J7R268 A0A067RGZ2 A0A1B6CL88 A0A1B6DWQ6 A0A069DXN6 A0A224XHU0 E0VKS8 A0A0P4VQV2 T1I0S2 A0A2P8ZP13 K7J1D5 A0A3L8E372 A0A026WG41 A0A1B6LZL6 E2BPI3 A0A1B6LYN0 A0A0J7L3T8 E2ARX0 D6WU14 A0A336MYR7 A0A336MMI0 U4URP0 Q16HF9 A0A0P6J6G8 A0A1S4G0N1 B3P4Z8 Q5BI37 Q9Y0W1 B4IJ38 A0A1W4VLZ3 Q9NG24 Q9V9T4 B4PP30 W4VS83 Q297V6 B4G333 A0A1I8MT49 B4LY73 B4N950 B0WSI2 A0A1Q3F0X5 B4K850 A0A0P5IMK9 A0A0P5KCC3 A0A1A9Y946 A0A1Q3F0Y3 A0A0P5J2K9 A0A1A9VYB7 A0A0P5EB74 A0A034VHK0 A0A0P5PFQ5 A0A0P5DVJ1 A0A0A1X5E9 A0A0P5QW73 A0A0P5IVG1 A0A0P4Z3I1 A0A1I8NLV6 A0A0P5Y798 A0A0P5RFM6 A0A0P5CAI9 A0A0P5YGS9 A0A0P5R5A9 A0A0N8CUS1 A0A0P5DBW9 A0A164PZN5 A0A1B6J014 A0A0P5ZUI0 A0A0N8DQW0 A0A0P5B560 A0A0P6EH49 A0A0P6DBZ2 A0A1B0B6S9 A0A0P5RHE4 A0A0P5TH33 A0A0P5HWE2 A0A3B0JHS7 A0A232ER24 A0A0P5Q6M3 A0A0P5YH39 A0A1B0AGK0 A0A0P6DM83 A0A0P6II66 A0A0P5RW57 A0A0C9PTD8 A0A0C9QM88 A0A0C9QVD7
A0A2J7R258 A0A2J7R251 A0A2J7R268 A0A067RGZ2 A0A1B6CL88 A0A1B6DWQ6 A0A069DXN6 A0A224XHU0 E0VKS8 A0A0P4VQV2 T1I0S2 A0A2P8ZP13 K7J1D5 A0A3L8E372 A0A026WG41 A0A1B6LZL6 E2BPI3 A0A1B6LYN0 A0A0J7L3T8 E2ARX0 D6WU14 A0A336MYR7 A0A336MMI0 U4URP0 Q16HF9 A0A0P6J6G8 A0A1S4G0N1 B3P4Z8 Q5BI37 Q9Y0W1 B4IJ38 A0A1W4VLZ3 Q9NG24 Q9V9T4 B4PP30 W4VS83 Q297V6 B4G333 A0A1I8MT49 B4LY73 B4N950 B0WSI2 A0A1Q3F0X5 B4K850 A0A0P5IMK9 A0A0P5KCC3 A0A1A9Y946 A0A1Q3F0Y3 A0A0P5J2K9 A0A1A9VYB7 A0A0P5EB74 A0A034VHK0 A0A0P5PFQ5 A0A0P5DVJ1 A0A0A1X5E9 A0A0P5QW73 A0A0P5IVG1 A0A0P4Z3I1 A0A1I8NLV6 A0A0P5Y798 A0A0P5RFM6 A0A0P5CAI9 A0A0P5YGS9 A0A0P5R5A9 A0A0N8CUS1 A0A0P5DBW9 A0A164PZN5 A0A1B6J014 A0A0P5ZUI0 A0A0N8DQW0 A0A0P5B560 A0A0P6EH49 A0A0P6DBZ2 A0A1B0B6S9 A0A0P5RHE4 A0A0P5TH33 A0A0P5HWE2 A0A3B0JHS7 A0A232ER24 A0A0P5Q6M3 A0A0P5YH39 A0A1B0AGK0 A0A0P6DM83 A0A0P6II66 A0A0P5RW57 A0A0C9PTD8 A0A0C9QM88 A0A0C9QVD7
Pubmed
19121390
22118469
26354079
24845553
26334808
20566863
+ More
27129103 29403074 20075255 30249741 24508170 20798317 18362917 19820115 23537049 17510324 26999592 17994087 10385622 11447119 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17550304 15632085 25315136 25348373 25830018 28648823
27129103 29403074 20075255 30249741 24508170 20798317 18362917 19820115 23537049 17510324 26999592 17994087 10385622 11447119 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17550304 15632085 25315136 25348373 25830018 28648823
EMBL
BABH01034235
BABH01034236
BABH01034237
NWSH01001542
PCG70911.1
ODYU01003717
+ More
SOQ42843.1 AGBW02011278 OWR46902.1 KQ459604 KPI92104.1 KQ460205 KPJ17020.1 NEVH01008200 PNF34920.1 PNF34918.1 PNF34919.1 KK852643 KDR19569.1 GEDC01023150 JAS14148.1 GEDC01007198 JAS30100.1 GBGD01000091 JAC88798.1 GFTR01008817 JAW07609.1 DS235250 EEB13984.1 GDKW01001189 JAI55406.1 ACPB03015533 PYGN01000005 PSN58246.1 QOIP01000001 RLU27190.1 KK107260 EZA54009.1 GEBQ01010848 JAT29129.1 GL449633 EFN82387.1 GEBQ01011263 JAT28714.1 LBMM01000985 KMQ97109.1 GL442192 EFN63819.1 KQ971352 EFA07691.2 UFQT01002451 SSX33497.1 UFQS01001380 UFQT01001380 SSX10606.1 SSX30289.1 KB632333 ERL92740.1 CH478175 EAT33689.1 GDUN01000079 JAN95840.1 CH954182 EDV52912.1 BT021387 AAX33535.1 AF148962 AAD38952.1 CH480846 EDW50991.1 AJ238397 CAB88669.1 AE014297 AAF57200.1 CM000160 EDW99266.1 GANO01000167 JAB59704.1 CM000070 EAL28099.2 CH479179 EDW24228.1 CH940650 EDW67961.1 CH964232 EDW81597.1 DS232072 EDS33912.1 GFDL01013838 JAV21207.1 CH933806 EDW15404.1 GDIQ01212086 JAK39639.1 GDIQ01212085 JAK39640.1 GFDL01013828 JAV21217.1 GDIQ01212084 JAK39641.1 GDIP01143916 JAJ79486.1 GAKP01017732 GAKP01017727 GAKP01017723 JAC41220.1 GDIQ01135216 JAL16510.1 GDIP01150170 JAJ73232.1 GBXI01008404 JAD05888.1 GDIQ01108919 JAL42807.1 GDIQ01215933 JAK35792.1 GDIP01230670 JAI92731.1 GDIP01061930 JAM41785.1 GDIQ01106617 JAL45109.1 GDIP01189509 JAJ33893.1 GDIP01057933 JAM45782.1 GDIQ01112462 JAL39264.1 GDIP01096879 JAM06836.1 GDIP01162317 JAJ61085.1 LRGB01002490 KZS07297.1 GECU01015207 JAS92499.1 GDIP01038385 JAM65330.1 GDIP01009767 JAM93948.1 GDIP01189510 JAJ33892.1 GDIQ01075531 JAN19206.1 GDIQ01079974 JAN14763.1 JXJN01009222 GDIQ01100685 JAL51041.1 GDIP01126321 JAL77393.1 GDIQ01222033 JAK29692.1 OUUW01000005 SPP80263.1 NNAY01002666 OXU20820.1 GDIQ01125125 JAL26601.1 GDIP01057934 JAM45781.1 GDIQ01075530 JAN19207.1 GDIQ01004021 JAN90716.1 GDIQ01100684 JAL51042.1 GBYB01004638 JAG74405.1 GBYB01004639 JAG74406.1 GBYB01004637 JAG74404.1
SOQ42843.1 AGBW02011278 OWR46902.1 KQ459604 KPI92104.1 KQ460205 KPJ17020.1 NEVH01008200 PNF34920.1 PNF34918.1 PNF34919.1 KK852643 KDR19569.1 GEDC01023150 JAS14148.1 GEDC01007198 JAS30100.1 GBGD01000091 JAC88798.1 GFTR01008817 JAW07609.1 DS235250 EEB13984.1 GDKW01001189 JAI55406.1 ACPB03015533 PYGN01000005 PSN58246.1 QOIP01000001 RLU27190.1 KK107260 EZA54009.1 GEBQ01010848 JAT29129.1 GL449633 EFN82387.1 GEBQ01011263 JAT28714.1 LBMM01000985 KMQ97109.1 GL442192 EFN63819.1 KQ971352 EFA07691.2 UFQT01002451 SSX33497.1 UFQS01001380 UFQT01001380 SSX10606.1 SSX30289.1 KB632333 ERL92740.1 CH478175 EAT33689.1 GDUN01000079 JAN95840.1 CH954182 EDV52912.1 BT021387 AAX33535.1 AF148962 AAD38952.1 CH480846 EDW50991.1 AJ238397 CAB88669.1 AE014297 AAF57200.1 CM000160 EDW99266.1 GANO01000167 JAB59704.1 CM000070 EAL28099.2 CH479179 EDW24228.1 CH940650 EDW67961.1 CH964232 EDW81597.1 DS232072 EDS33912.1 GFDL01013838 JAV21207.1 CH933806 EDW15404.1 GDIQ01212086 JAK39639.1 GDIQ01212085 JAK39640.1 GFDL01013828 JAV21217.1 GDIQ01212084 JAK39641.1 GDIP01143916 JAJ79486.1 GAKP01017732 GAKP01017727 GAKP01017723 JAC41220.1 GDIQ01135216 JAL16510.1 GDIP01150170 JAJ73232.1 GBXI01008404 JAD05888.1 GDIQ01108919 JAL42807.1 GDIQ01215933 JAK35792.1 GDIP01230670 JAI92731.1 GDIP01061930 JAM41785.1 GDIQ01106617 JAL45109.1 GDIP01189509 JAJ33893.1 GDIP01057933 JAM45782.1 GDIQ01112462 JAL39264.1 GDIP01096879 JAM06836.1 GDIP01162317 JAJ61085.1 LRGB01002490 KZS07297.1 GECU01015207 JAS92499.1 GDIP01038385 JAM65330.1 GDIP01009767 JAM93948.1 GDIP01189510 JAJ33892.1 GDIQ01075531 JAN19206.1 GDIQ01079974 JAN14763.1 JXJN01009222 GDIQ01100685 JAL51041.1 GDIP01126321 JAL77393.1 GDIQ01222033 JAK29692.1 OUUW01000005 SPP80263.1 NNAY01002666 OXU20820.1 GDIQ01125125 JAL26601.1 GDIP01057934 JAM45781.1 GDIQ01075530 JAN19207.1 GDIQ01004021 JAN90716.1 GDIQ01100684 JAL51042.1 GBYB01004638 JAG74405.1 GBYB01004639 JAG74406.1 GBYB01004637 JAG74404.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053268
UP000053240
UP000235965
+ More
UP000027135 UP000009046 UP000015103 UP000245037 UP000002358 UP000279307 UP000053097 UP000008237 UP000036403 UP000000311 UP000007266 UP000030742 UP000008820 UP000008711 UP000001292 UP000192221 UP000000803 UP000002282 UP000001819 UP000008744 UP000095301 UP000008792 UP000007798 UP000002320 UP000009192 UP000092443 UP000078200 UP000095300 UP000076858 UP000092460 UP000268350 UP000215335 UP000092445
UP000027135 UP000009046 UP000015103 UP000245037 UP000002358 UP000279307 UP000053097 UP000008237 UP000036403 UP000000311 UP000007266 UP000030742 UP000008820 UP000008711 UP000001292 UP000192221 UP000000803 UP000002282 UP000001819 UP000008744 UP000095301 UP000008792 UP000007798 UP000002320 UP000009192 UP000092443 UP000078200 UP000095300 UP000076858 UP000092460 UP000268350 UP000215335 UP000092445
Pfam
Interpro
IPR019787
Znf_PHD-finger
+ More
IPR011011 Znf_FYVE_PHD
IPR013136 WSTF_Acf1_Cbp146
IPR018501 DDT_dom
IPR028941 WHIM2_dom
IPR028942 WHIM1_dom
IPR001487 Bromodomain
IPR013083 Znf_RING/FYVE/PHD
IPR001965 Znf_PHD
IPR036427 Bromodomain-like_sf
IPR019786 Zinc_finger_PHD-type_CS
IPR018359 Bromodomain_CS
IPR037325 Acf1_Bromo
IPR001841 Znf_RING
IPR011011 Znf_FYVE_PHD
IPR013136 WSTF_Acf1_Cbp146
IPR018501 DDT_dom
IPR028941 WHIM2_dom
IPR028942 WHIM1_dom
IPR001487 Bromodomain
IPR013083 Znf_RING/FYVE/PHD
IPR001965 Znf_PHD
IPR036427 Bromodomain-like_sf
IPR019786 Zinc_finger_PHD-type_CS
IPR018359 Bromodomain_CS
IPR037325 Acf1_Bromo
IPR001841 Znf_RING
Gene 3D
ProteinModelPortal
H9IU84
A0A2A4JG07
A0A2H1VPS5
A0A212EZJ2
A0A194PHB9
A0A194RI66
+ More
A0A2J7R258 A0A2J7R251 A0A2J7R268 A0A067RGZ2 A0A1B6CL88 A0A1B6DWQ6 A0A069DXN6 A0A224XHU0 E0VKS8 A0A0P4VQV2 T1I0S2 A0A2P8ZP13 K7J1D5 A0A3L8E372 A0A026WG41 A0A1B6LZL6 E2BPI3 A0A1B6LYN0 A0A0J7L3T8 E2ARX0 D6WU14 A0A336MYR7 A0A336MMI0 U4URP0 Q16HF9 A0A0P6J6G8 A0A1S4G0N1 B3P4Z8 Q5BI37 Q9Y0W1 B4IJ38 A0A1W4VLZ3 Q9NG24 Q9V9T4 B4PP30 W4VS83 Q297V6 B4G333 A0A1I8MT49 B4LY73 B4N950 B0WSI2 A0A1Q3F0X5 B4K850 A0A0P5IMK9 A0A0P5KCC3 A0A1A9Y946 A0A1Q3F0Y3 A0A0P5J2K9 A0A1A9VYB7 A0A0P5EB74 A0A034VHK0 A0A0P5PFQ5 A0A0P5DVJ1 A0A0A1X5E9 A0A0P5QW73 A0A0P5IVG1 A0A0P4Z3I1 A0A1I8NLV6 A0A0P5Y798 A0A0P5RFM6 A0A0P5CAI9 A0A0P5YGS9 A0A0P5R5A9 A0A0N8CUS1 A0A0P5DBW9 A0A164PZN5 A0A1B6J014 A0A0P5ZUI0 A0A0N8DQW0 A0A0P5B560 A0A0P6EH49 A0A0P6DBZ2 A0A1B0B6S9 A0A0P5RHE4 A0A0P5TH33 A0A0P5HWE2 A0A3B0JHS7 A0A232ER24 A0A0P5Q6M3 A0A0P5YH39 A0A1B0AGK0 A0A0P6DM83 A0A0P6II66 A0A0P5RW57 A0A0C9PTD8 A0A0C9QM88 A0A0C9QVD7
A0A2J7R258 A0A2J7R251 A0A2J7R268 A0A067RGZ2 A0A1B6CL88 A0A1B6DWQ6 A0A069DXN6 A0A224XHU0 E0VKS8 A0A0P4VQV2 T1I0S2 A0A2P8ZP13 K7J1D5 A0A3L8E372 A0A026WG41 A0A1B6LZL6 E2BPI3 A0A1B6LYN0 A0A0J7L3T8 E2ARX0 D6WU14 A0A336MYR7 A0A336MMI0 U4URP0 Q16HF9 A0A0P6J6G8 A0A1S4G0N1 B3P4Z8 Q5BI37 Q9Y0W1 B4IJ38 A0A1W4VLZ3 Q9NG24 Q9V9T4 B4PP30 W4VS83 Q297V6 B4G333 A0A1I8MT49 B4LY73 B4N950 B0WSI2 A0A1Q3F0X5 B4K850 A0A0P5IMK9 A0A0P5KCC3 A0A1A9Y946 A0A1Q3F0Y3 A0A0P5J2K9 A0A1A9VYB7 A0A0P5EB74 A0A034VHK0 A0A0P5PFQ5 A0A0P5DVJ1 A0A0A1X5E9 A0A0P5QW73 A0A0P5IVG1 A0A0P4Z3I1 A0A1I8NLV6 A0A0P5Y798 A0A0P5RFM6 A0A0P5CAI9 A0A0P5YGS9 A0A0P5R5A9 A0A0N8CUS1 A0A0P5DBW9 A0A164PZN5 A0A1B6J014 A0A0P5ZUI0 A0A0N8DQW0 A0A0P5B560 A0A0P6EH49 A0A0P6DBZ2 A0A1B0B6S9 A0A0P5RHE4 A0A0P5TH33 A0A0P5HWE2 A0A3B0JHS7 A0A232ER24 A0A0P5Q6M3 A0A0P5YH39 A0A1B0AGK0 A0A0P6DM83 A0A0P6II66 A0A0P5RW57 A0A0C9PTD8 A0A0C9QM88 A0A0C9QVD7
PDB
4TVR
E-value=0.000492054,
Score=105
Ontologies
GO
GO:0005634
GO:0046872
GO:0016021
GO:0006338
GO:0046020
GO:0006348
GO:0000781
GO:0007399
GO:0043462
GO:0006334
GO:0016590
GO:0031445
GO:0042766
GO:0016584
GO:0003677
GO:0045892
GO:0008623
GO:0070615
GO:0006355
GO:0016573
GO:0031497
GO:0006333
GO:0016746
GO:0005515
GO:0003899
GO:0006351
GO:0006886
GO:0016192
GO:0030117
GO:0007154
GO:0016020
GO:0005112
GO:0015930
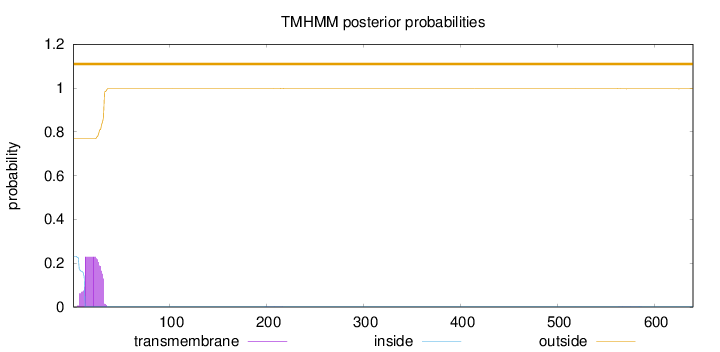
Topology
Subcellular location
Nucleus
Length:
640
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
4.72867999999998
Exp number, first 60 AAs:
4.69742
Total prob of N-in:
0.23140
outside
1 - 640
Population Genetic Test Statistics
Pi
171.757705
Theta
133.247582
Tajima's D
0.888118
CLR
0.453951
CSRT
0.626618669066547
Interpretation
Uncertain
