Pre Gene Modal
BGIBMGA000813
Annotation
PREDICTED:_retinol_dehydrogenase_13-like_[Papilio_polytes]
Location in the cell
Cytoplasmic Reliability : 1.05 Mitochondrial Reliability : 1.278
Sequence
CDS
ATGTGGGTTCCCAGCACGCCGGTAACTTTGGTGACTGCAGTAGCGGCCGTGGCTGGTGGGGTGTGCATATTCAAAGATTACTACAGTGGTCCTCCGTACGACCGGTCCGTCCGTGCCGACAACAGAACAATTATAATTACTGGTGCTAACAGCGGTATCGGGAAAGCTGCAGCAAAAGACTTCGCCAAAAGGGGTGCTAAGGTGTTTATGGCGTGCCGTGACGTCAAGAAGTGCGAGGAGCAACGACGCGACATCGTGCTTGAGACGAACAACAAGTACGTGTACTGCAGACCCTGCGACCTGGCCAGCACGGCCTCCATCAGAGAGTTCGTTGCCAGATTCAAAGGAGAGGAGCCACACCTTCATGTGCTGGTCAATAACGCCGGGGTGATGGAACCACCCAAGGGAGTCACTAAGGACGGCTTCGAGACCCAGCTCGGGGTCAATCACCTCGGACATTTCTTGCTGACAAACTTGCTCCTGGATACGCTCAAGGCCTCAGCCCCGAGCCGCGTGGTGGTGGTGTCGAGCAGCGCGCACCAGCGCGGGCGGATCCACAAGCAGGACCTCAACATGAGCGAGAACTATGACGCGTCCGCCGCCTACAGCCAGAGCAAGCTCGCCAACCTACTGTTCGCGCGGGAGCTCGGCAGGAGGTTACTCGGAACTGGAGTGGCGACAATCGCAGTAGACCCTGGCCTCGTAGACACGGACATCACCCGCCACATGTCAATGATGAAAAGCGTTACTCGGTTCTTCATATACCCACTGTTCTGGCCCTTCATGAAGAAACCCGAAATCGGAGGGCAAACAGTAGTACACGCTGCCCTAGACCCTAAGCTGAAAGACTGCGCCGGAGACTATTACGTTGATATGAAGAAAGCTGAACCATCACAAGAAGCTCAAGACTTCGAGCTGGCTCAGTGGCTGTGGAGGGTCAGCGAGGCGTGGACCAAGCTGGACGAGCATAAACAAGCCATCGCCAAAGACACACGGAATTAG
Protein
MWVPSTPVTLVTAVAAVAGGVCIFKDYYSGPPYDRSVRADNRTIIITGANSGIGKAAAKDFAKRGAKVFMACRDVKKCEEQRRDIVLETNNKYVYCRPCDLASTASIREFVARFKGEEPHLHVLVNNAGVMEPPKGVTKDGFETQLGVNHLGHFLLTNLLLDTLKASAPSRVVVVSSSAHQRGRIHKQDLNMSENYDASAAYSQSKLANLLFARELGRRLLGTGVATIAVDPGLVDTDITRHMSMMKSVTRFFIYPLFWPFMKKPEIGGQTVVHAALDPKLKDCAGDYYVDMKKAEPSQEAQDFELAQWLWRVSEAWTKLDEHKQAIAKDTRN
Summary
Similarity
Belongs to the short-chain dehydrogenases/reductases (SDR) family.
Uniprot
H9IU83
A0A2W1BPB9
A0A2H1V0I4
A0A2A4JGG3
A0A3S2NRJ6
A0A194PFG2
+ More
A0A194RGS8 A0A1W4WYE7 A0A1B6H9T6 T1I4Q4 A0A1B0AAX4 A0A1A9UWG7 A0A1B6EUT9 A0A1B0AYR4 E9J8J2 A0A0T6BDA0 A0A224XTS0 A0A1A9YGG4 A0A195FCN8 A0A1B0FE94 A0A067RLP9 E2C6J3 A0A1B6LTS6 B3N9W1 A0A158NBV6 A0A0K8TLB0 A0A1I8M9J0 A0A2M4BTT4 B4J7Z2 B4H8I4 A0A2M3ZCQ8 A0A1W4VZF7 A0A0M4EVS0 W8CDP7 Q28YK4 A0A084W0X3 W5JL96 A0A1A9W9N4 F4X8E8 A0A151X9L1 A0A2M4BTU8 A0A2M4AW80 A0A195BEY3 A0A0J9R7K6 B4HR47 Q7JUS1 B4P1S7 B4QEQ2 A0A182TVW4 A0A182YCZ5 W8C5R8 A0A0J7KBW3 A0A151J924 A0A0L0C2H0 A0A0Q9W4U8 A0A182WKM2 U5EU75 A0A182TCC6 A0A182UVS1 A0A1C7CZE8 A0A182I1T9 A0A182X743 B0WVA8 Q7Q686 A0A1L8EH13 A0A3B0J367 A0A0M9A417 B4N649 A0A0R3NU57 B3MHR9 A0A182FGX8 A0A0Q9WH74 K7IUK5 A0A182K5N8 A0A182P5J6 A0A3L8DTE9 E2AR14 A0A1Q3FGF3 A0A232F458 A0A158NBV5 A0A1Q3FGI4 A0A1S4F244 A0A1Y1JX22 A0A1L8E4K6 A0A195C9Z0 A0A182REG7 A0A182NIK0 A0A1I8PH20 A0A2A3ERX9 A0A182MJR3 A0A087ZZP4 A0A336LUI6 A0A1B6C7X2 A0A182H592 A0A0K8VTD1 Q17I51 A0A0A1X931 A0A034W0F6 A0A026X3N1 N6SYJ7
A0A194RGS8 A0A1W4WYE7 A0A1B6H9T6 T1I4Q4 A0A1B0AAX4 A0A1A9UWG7 A0A1B6EUT9 A0A1B0AYR4 E9J8J2 A0A0T6BDA0 A0A224XTS0 A0A1A9YGG4 A0A195FCN8 A0A1B0FE94 A0A067RLP9 E2C6J3 A0A1B6LTS6 B3N9W1 A0A158NBV6 A0A0K8TLB0 A0A1I8M9J0 A0A2M4BTT4 B4J7Z2 B4H8I4 A0A2M3ZCQ8 A0A1W4VZF7 A0A0M4EVS0 W8CDP7 Q28YK4 A0A084W0X3 W5JL96 A0A1A9W9N4 F4X8E8 A0A151X9L1 A0A2M4BTU8 A0A2M4AW80 A0A195BEY3 A0A0J9R7K6 B4HR47 Q7JUS1 B4P1S7 B4QEQ2 A0A182TVW4 A0A182YCZ5 W8C5R8 A0A0J7KBW3 A0A151J924 A0A0L0C2H0 A0A0Q9W4U8 A0A182WKM2 U5EU75 A0A182TCC6 A0A182UVS1 A0A1C7CZE8 A0A182I1T9 A0A182X743 B0WVA8 Q7Q686 A0A1L8EH13 A0A3B0J367 A0A0M9A417 B4N649 A0A0R3NU57 B3MHR9 A0A182FGX8 A0A0Q9WH74 K7IUK5 A0A182K5N8 A0A182P5J6 A0A3L8DTE9 E2AR14 A0A1Q3FGF3 A0A232F458 A0A158NBV5 A0A1Q3FGI4 A0A1S4F244 A0A1Y1JX22 A0A1L8E4K6 A0A195C9Z0 A0A182REG7 A0A182NIK0 A0A1I8PH20 A0A2A3ERX9 A0A182MJR3 A0A087ZZP4 A0A336LUI6 A0A1B6C7X2 A0A182H592 A0A0K8VTD1 Q17I51 A0A0A1X931 A0A034W0F6 A0A026X3N1 N6SYJ7
Pubmed
19121390
28756777
26354079
21282665
24845553
20798317
+ More
17994087 21347285 26369729 25315136 24495485 15632085 24438588 20920257 23761445 21719571 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 25244985 26108605 20966253 12364791 14747013 17210077 20075255 30249741 28648823 28004739 26483478 17510324 25830018 25348373 24508170 23537049
17994087 21347285 26369729 25315136 24495485 15632085 24438588 20920257 23761445 21719571 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 25244985 26108605 20966253 12364791 14747013 17210077 20075255 30249741 28648823 28004739 26483478 17510324 25830018 25348373 24508170 23537049
EMBL
BABH01034229
BABH01034230
KZ149945
PZC76818.1
ODYU01000125
SOQ34373.1
+ More
NWSH01001542 PCG70909.1 RSAL01000004 RVE54553.1 KQ459604 KPI92106.1 KQ460205 KPJ17023.1 GECU01036314 JAS71392.1 ACPB03014599 GECZ01028004 JAS41765.1 JXJN01005924 GL769034 EFZ10865.1 LJIG01001628 KRT85312.1 GFTR01005075 JAW11351.1 KQ981673 KYN38405.1 CCAG010000559 KK852433 KDR23963.1 GL453160 EFN76395.1 GEBQ01012958 JAT27019.1 CH954177 EDV59657.1 ADTU01011331 GDAI01002635 JAI14968.1 GGFJ01007283 MBW56424.1 CH916367 EDW02222.1 CH479223 EDW35019.1 GGFM01005489 MBW26240.1 CP012524 ALC42025.1 GAMC01001576 JAC04980.1 CM000071 EAL25962.2 ATLV01019163 KE525263 KFB43867.1 ADMH02001240 ETN63539.1 GL888932 EGI57150.1 KQ982373 KYQ57052.1 GGFJ01007282 MBW56423.1 GGFK01011723 MBW45044.1 KQ976500 KYM83151.1 CM002911 KMY92051.1 CH480816 EDW46790.1 AE013599 AY122067 AAF59216.3 AAM52579.1 CM000157 EDW89213.1 CM000362 EDX06048.1 GAMC01001577 JAC04979.1 LBMM01009835 KMQ87853.1 KQ979433 KYN21565.1 JRES01000975 KNC26553.1 CH940648 KRF80051.1 GANO01002446 JAB57425.1 APCN01000093 DS232121 EDS35451.1 AAAB01008960 EAA10915.4 GFDG01000855 JAV17944.1 OUUW01000001 SPP75735.1 KQ435741 KOX76785.1 CH964154 EDW79838.2 KRT02701.1 CH902619 EDV36906.1 KRF80053.1 AAZX01004215 QOIP01000004 RLU23572.1 GL441846 EFN64160.1 GFDL01008421 JAV26624.1 NNAY01001089 OXU25177.1 GFDL01008390 JAV26655.1 GEZM01103119 JAV51556.1 GFDF01000414 JAV13670.1 KQ978068 KYM97530.1 KZ288191 PBC34450.1 AXCM01002675 UFQS01000102 UFQT01000102 SSW99637.1 SSX20017.1 GEDC01027702 JAS09596.1 JXUM01025925 KQ560738 KXJ81071.1 GDHF01010201 GDHF01002729 JAI42113.1 JAI49585.1 CH477243 EAT46307.1 GBXI01007042 JAD07250.1 GAKP01011739 JAC47213.1 KK107036 EZA62004.1 APGK01059446 KB741293 ENN70283.1
NWSH01001542 PCG70909.1 RSAL01000004 RVE54553.1 KQ459604 KPI92106.1 KQ460205 KPJ17023.1 GECU01036314 JAS71392.1 ACPB03014599 GECZ01028004 JAS41765.1 JXJN01005924 GL769034 EFZ10865.1 LJIG01001628 KRT85312.1 GFTR01005075 JAW11351.1 KQ981673 KYN38405.1 CCAG010000559 KK852433 KDR23963.1 GL453160 EFN76395.1 GEBQ01012958 JAT27019.1 CH954177 EDV59657.1 ADTU01011331 GDAI01002635 JAI14968.1 GGFJ01007283 MBW56424.1 CH916367 EDW02222.1 CH479223 EDW35019.1 GGFM01005489 MBW26240.1 CP012524 ALC42025.1 GAMC01001576 JAC04980.1 CM000071 EAL25962.2 ATLV01019163 KE525263 KFB43867.1 ADMH02001240 ETN63539.1 GL888932 EGI57150.1 KQ982373 KYQ57052.1 GGFJ01007282 MBW56423.1 GGFK01011723 MBW45044.1 KQ976500 KYM83151.1 CM002911 KMY92051.1 CH480816 EDW46790.1 AE013599 AY122067 AAF59216.3 AAM52579.1 CM000157 EDW89213.1 CM000362 EDX06048.1 GAMC01001577 JAC04979.1 LBMM01009835 KMQ87853.1 KQ979433 KYN21565.1 JRES01000975 KNC26553.1 CH940648 KRF80051.1 GANO01002446 JAB57425.1 APCN01000093 DS232121 EDS35451.1 AAAB01008960 EAA10915.4 GFDG01000855 JAV17944.1 OUUW01000001 SPP75735.1 KQ435741 KOX76785.1 CH964154 EDW79838.2 KRT02701.1 CH902619 EDV36906.1 KRF80053.1 AAZX01004215 QOIP01000004 RLU23572.1 GL441846 EFN64160.1 GFDL01008421 JAV26624.1 NNAY01001089 OXU25177.1 GFDL01008390 JAV26655.1 GEZM01103119 JAV51556.1 GFDF01000414 JAV13670.1 KQ978068 KYM97530.1 KZ288191 PBC34450.1 AXCM01002675 UFQS01000102 UFQT01000102 SSW99637.1 SSX20017.1 GEDC01027702 JAS09596.1 JXUM01025925 KQ560738 KXJ81071.1 GDHF01010201 GDHF01002729 JAI42113.1 JAI49585.1 CH477243 EAT46307.1 GBXI01007042 JAD07250.1 GAKP01011739 JAC47213.1 KK107036 EZA62004.1 APGK01059446 KB741293 ENN70283.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000053268
UP000053240
UP000192223
+ More
UP000015103 UP000092445 UP000078200 UP000092460 UP000092443 UP000078541 UP000092444 UP000027135 UP000008237 UP000008711 UP000005205 UP000095301 UP000001070 UP000008744 UP000192221 UP000092553 UP000001819 UP000030765 UP000000673 UP000091820 UP000007755 UP000075809 UP000078540 UP000001292 UP000000803 UP000002282 UP000000304 UP000075902 UP000076408 UP000036403 UP000078492 UP000037069 UP000008792 UP000075920 UP000075901 UP000075903 UP000075882 UP000075840 UP000076407 UP000002320 UP000007062 UP000268350 UP000053105 UP000007798 UP000007801 UP000069272 UP000002358 UP000075881 UP000075885 UP000279307 UP000000311 UP000215335 UP000078542 UP000075900 UP000075884 UP000095300 UP000242457 UP000075883 UP000005203 UP000069940 UP000249989 UP000008820 UP000053097 UP000019118
UP000015103 UP000092445 UP000078200 UP000092460 UP000092443 UP000078541 UP000092444 UP000027135 UP000008237 UP000008711 UP000005205 UP000095301 UP000001070 UP000008744 UP000192221 UP000092553 UP000001819 UP000030765 UP000000673 UP000091820 UP000007755 UP000075809 UP000078540 UP000001292 UP000000803 UP000002282 UP000000304 UP000075902 UP000076408 UP000036403 UP000078492 UP000037069 UP000008792 UP000075920 UP000075901 UP000075903 UP000075882 UP000075840 UP000076407 UP000002320 UP000007062 UP000268350 UP000053105 UP000007798 UP000007801 UP000069272 UP000002358 UP000075881 UP000075885 UP000279307 UP000000311 UP000215335 UP000078542 UP000075900 UP000075884 UP000095300 UP000242457 UP000075883 UP000005203 UP000069940 UP000249989 UP000008820 UP000053097 UP000019118
Interpro
Gene 3D
ProteinModelPortal
H9IU83
A0A2W1BPB9
A0A2H1V0I4
A0A2A4JGG3
A0A3S2NRJ6
A0A194PFG2
+ More
A0A194RGS8 A0A1W4WYE7 A0A1B6H9T6 T1I4Q4 A0A1B0AAX4 A0A1A9UWG7 A0A1B6EUT9 A0A1B0AYR4 E9J8J2 A0A0T6BDA0 A0A224XTS0 A0A1A9YGG4 A0A195FCN8 A0A1B0FE94 A0A067RLP9 E2C6J3 A0A1B6LTS6 B3N9W1 A0A158NBV6 A0A0K8TLB0 A0A1I8M9J0 A0A2M4BTT4 B4J7Z2 B4H8I4 A0A2M3ZCQ8 A0A1W4VZF7 A0A0M4EVS0 W8CDP7 Q28YK4 A0A084W0X3 W5JL96 A0A1A9W9N4 F4X8E8 A0A151X9L1 A0A2M4BTU8 A0A2M4AW80 A0A195BEY3 A0A0J9R7K6 B4HR47 Q7JUS1 B4P1S7 B4QEQ2 A0A182TVW4 A0A182YCZ5 W8C5R8 A0A0J7KBW3 A0A151J924 A0A0L0C2H0 A0A0Q9W4U8 A0A182WKM2 U5EU75 A0A182TCC6 A0A182UVS1 A0A1C7CZE8 A0A182I1T9 A0A182X743 B0WVA8 Q7Q686 A0A1L8EH13 A0A3B0J367 A0A0M9A417 B4N649 A0A0R3NU57 B3MHR9 A0A182FGX8 A0A0Q9WH74 K7IUK5 A0A182K5N8 A0A182P5J6 A0A3L8DTE9 E2AR14 A0A1Q3FGF3 A0A232F458 A0A158NBV5 A0A1Q3FGI4 A0A1S4F244 A0A1Y1JX22 A0A1L8E4K6 A0A195C9Z0 A0A182REG7 A0A182NIK0 A0A1I8PH20 A0A2A3ERX9 A0A182MJR3 A0A087ZZP4 A0A336LUI6 A0A1B6C7X2 A0A182H592 A0A0K8VTD1 Q17I51 A0A0A1X931 A0A034W0F6 A0A026X3N1 N6SYJ7
A0A194RGS8 A0A1W4WYE7 A0A1B6H9T6 T1I4Q4 A0A1B0AAX4 A0A1A9UWG7 A0A1B6EUT9 A0A1B0AYR4 E9J8J2 A0A0T6BDA0 A0A224XTS0 A0A1A9YGG4 A0A195FCN8 A0A1B0FE94 A0A067RLP9 E2C6J3 A0A1B6LTS6 B3N9W1 A0A158NBV6 A0A0K8TLB0 A0A1I8M9J0 A0A2M4BTT4 B4J7Z2 B4H8I4 A0A2M3ZCQ8 A0A1W4VZF7 A0A0M4EVS0 W8CDP7 Q28YK4 A0A084W0X3 W5JL96 A0A1A9W9N4 F4X8E8 A0A151X9L1 A0A2M4BTU8 A0A2M4AW80 A0A195BEY3 A0A0J9R7K6 B4HR47 Q7JUS1 B4P1S7 B4QEQ2 A0A182TVW4 A0A182YCZ5 W8C5R8 A0A0J7KBW3 A0A151J924 A0A0L0C2H0 A0A0Q9W4U8 A0A182WKM2 U5EU75 A0A182TCC6 A0A182UVS1 A0A1C7CZE8 A0A182I1T9 A0A182X743 B0WVA8 Q7Q686 A0A1L8EH13 A0A3B0J367 A0A0M9A417 B4N649 A0A0R3NU57 B3MHR9 A0A182FGX8 A0A0Q9WH74 K7IUK5 A0A182K5N8 A0A182P5J6 A0A3L8DTE9 E2AR14 A0A1Q3FGF3 A0A232F458 A0A158NBV5 A0A1Q3FGI4 A0A1S4F244 A0A1Y1JX22 A0A1L8E4K6 A0A195C9Z0 A0A182REG7 A0A182NIK0 A0A1I8PH20 A0A2A3ERX9 A0A182MJR3 A0A087ZZP4 A0A336LUI6 A0A1B6C7X2 A0A182H592 A0A0K8VTD1 Q17I51 A0A0A1X931 A0A034W0F6 A0A026X3N1 N6SYJ7
PDB
3RD5
E-value=7.79997e-29,
Score=316
Ontologies
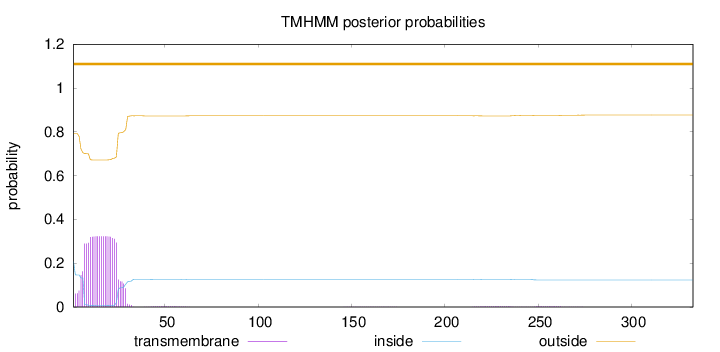
Topology
Length:
333
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
6.90627
Exp number, first 60 AAs:
6.78796
Total prob of N-in:
0.20817
outside
1 - 333
Population Genetic Test Statistics
Pi
175.093735
Theta
160.219726
Tajima's D
0.541423
CLR
0.786309
CSRT
0.531073446327684
Interpretation
Uncertain
