Gene
KWMTBOMO14951
Pre Gene Modal
BGIBMGA000812
Annotation
PREDICTED:_patj_homolog_[Papilio_machaon]
Full name
Patj homolog
Location in the cell
Cytoplasmic Reliability : 1.969
Sequence
CDS
ATGGTTCTAAGTACCGAATGGGCACAAGTTGAAGTTGTCGAGCTCACTAATGATGGCAACGGCTTGGGTTTCAACCTAGTCGGCGGCAGGAGTACCGGCGTCGTGATAAAATACGTACTGCCAGGAGGTGTGGCTGACAAGGACAGGCGACTGCAAAGCGGTGATCACGTGCTGCAGGTGGGCTCCGTAAACCTGCGCGGCTTCACATCGGAGCAGGTGGCTGCTGTATTGAGGCAGGCGGGACCAGTAGTCCGTTTGTTAGTCGCGCGCCCAGCGGATCCTGCGACCGCTCTCCGTGCCCCGCAACCCGGCACAGCGCTCGTGCCGACCAAAATCCTTGCTGATCCGGAGCTTCTCGACCGTCATCTCATCGAAACTGGTTACGGTGCAGTGTATGACTTATCTCAATGTTACAGTGACTATAATATCAATGGGCAAAATGGCGAAACGGAAACTAGTATAGTGGCAGCAGCGGTTAGTGTTATTGGTGACAATCCACAGAAAATACCTGATCATCCCATTGTATGTGATTCTGTGTCACCAACCATCACTATCACTGTTCCAGTAGAGTTACCTGAAATTGAAATTATTCATGTGGATTTAAATAAGAATGTGTATGGACTAGGTATCACGGTAGCGGGGTATGTGTGTGAAAAAGAGGAGCTCTCTGGAATATTTGTTAAAAGTATAATCGAGGGTAGCAGTGCTGAACAAAGTGGTAAAATTAAAATTAATGATAGAATTATTGAAGTGGATGGTATATCATTAGCCGATAAGAGTAATCCCCAGTCTGTGGAGATATTGAGGAACACAGGTATTTCTGTTCACTTAGTTTTAGAGAGATATCTTCGAGGTCCAAAATTTGAACATCTACAACTAGCTATATTTAACGAGGAGCGGCCAACTTCACCATCACCCTCCACCACTACACTTTCTTGGTTCCCAGTGCCTTCTCAACTCGAAAATAGTATAACAGAAATAGAACCTGAACCAGAATCAAATACTACAATTGATTCCAGCGTCTTAGAGGTGGGAGATCTTCAGGAGCCCACACAAGAAGAGCTGGACCAAAGATTTGATGAAATACTTGCTGTTGACAAAGAGGACATTGTAAAGAAGTGGAAAGAAGAAATTGGATCTGACAAAGAAATATTAGTGGCTGAAATAAAAAAGTTAGAAGGACTAGGGATCAGCCTTGAAGGAACTGTGGATGTGGAAGGAGGGCAAGAGTTGAGACCTCATCACTACATTAGGTCTGTGCTTCCAGATGGGCCTGTTGGACTGCAGGGTGTACTAAATGCTGGTGATGAACTGCTCGAAGTCAATGAATATAGATTACATGGCTTAACACATTCGGAAGTTGTAAACATTCTAAAGCAACTACCAGACAGAGTAAGATTGGTATGTGCACGTAACAGCGTCGACAGTGGACCACGCCCTGTTGTAAACTTAGCATCAGATCGAGAAGGGTTTGAAGCGAGGAAAATTATATCCGGAAGCCTAAACAACCTCACAACATTAGTAAAAGCACAATCAGATACATCTATAAACACATCAAGTACAGCAACACTGACTAATCACTCAGAGCAGTCTAAGAAATCCCGATCTCTTGAGTGTGTGTCTGGTTTAGCCATGTGGCAGAGCAAAGAGGATATTGTGATGCTTGTCAAAGCTGATCAGGGACTCGGCTTTTCAATATTAGATTACCAAGATCCAGTAGATCCAAATGGAACAGTCATTGTCGTTAGGAGTTTAGTACCCGGCGGAGTTGCAGAGAAGAATGGTGAAATATCACCAGGAGACAGGGTAATGTCCGTGAACGGAGTCAGCATACAAAATGCGACATTAGACCAAGCTGTTCAGGCTCTCAAAGGTGCTCCGAGGGGGACAGTCAAAGTAGGAATTGCGAGACCCTTGCCACCTCAAGACTCCTCAAAGTCCAAGGGTACAATTAATGTTAACAAAGCAAGCTGA
Protein
MVLSTEWAQVEVVELTNDGNGLGFNLVGGRSTGVVIKYVLPGGVADKDRRLQSGDHVLQVGSVNLRGFTSEQVAAVLRQAGPVVRLLVARPADPATALRAPQPGTALVPTKILADPELLDRHLIETGYGAVYDLSQCYSDYNINGQNGETETSIVAAAVSVIGDNPQKIPDHPIVCDSVSPTITITVPVELPEIEIIHVDLNKNVYGLGITVAGYVCEKEELSGIFVKSIIEGSSAEQSGKIKINDRIIEVDGISLADKSNPQSVEILRNTGISVHLVLERYLRGPKFEHLQLAIFNEERPTSPSPSTTTLSWFPVPSQLENSITEIEPEPESNTTIDSSVLEVGDLQEPTQEELDQRFDEILAVDKEDIVKKWKEEIGSDKEILVAEIKKLEGLGISLEGTVDVEGGQELRPHHYIRSVLPDGPVGLQGVLNAGDELLEVNEYRLHGLTHSEVVNILKQLPDRVRLVCARNSVDSGPRPVVNLASDREGFEARKIISGSLNNLTTLVKAQSDTSINTSSTATLTNHSEQSKKSRSLECVSGLAMWQSKEDIVMLVKADQGLGFSILDYQDPVDPNGTVIVVRSLVPGGVAEKNGEISPGDRVMSVNGVSIQNATLDQAVQALKGAPRGTVKVGIARPLPPQDSSKSKGTINVNKAS
Summary
Description
Involved in cell polarity establishment. Probably participates in the assembly, positioning and maintenance of adherens junctions via its interaction with the SAC complex.
Subunit
Component of the SAC complex, a complex composed of crb, Patj and sdt. Interacts directly with nrx via its third and fourth PDZ domains. Interacts directly with par-6, possibly mediating a link between the SAC complex and the par-6 complex, which is composed of par-6, baz and aPKC.
Similarity
Belongs to the Patj family.
Keywords
Complete proteome
Developmental protein
Membrane
Phosphoprotein
Reference proteome
Repeat
Feature
chain Patj homolog
Uniprot
H9IU82
A0A2A4JGR2
A0A2H1V279
A0A2W1BUE1
A0A194RHV9
A0A194PFP2
+ More
A0A212ERH9 A0A0L7KSF9 E2B674 A0A0M8ZVG3 A0A158NCF0 F4WWZ8 A0A154PNZ1 A0A026X228 A0A195BWA7 A0A151JQU8 A0A1Y1M2Z1 A0A0L7R1E0 A0A2J7R5D1 A0A2J7R5D6 A0A2J7R5D2 A0A2J7R5D3 A0A1B6GQS6 A0A1B6HFI9 A0A1B6HTA6 A0A1B6ELN7 A0A087ZV81 D6WRN8 A0A151WMY2 A0A139WEE0 D6WPR0 A0A0P4VIU4 A0A146KZ93 A0A232FA68 A0A023F3W4 A0A224X4S0 A0A146LAK9 A0A151IBX1 E0VGE8 A0A3L8DGH0 A0A0A9YHT4 A0A182GIF1 A0A336KTG0 A0A1B0DAI8 A0A1D2MTJ7 B0WHT5 B4PDU5 A0A1Y1MNC1 A0A0J9RM08 D6W6M5 B3NEV8 A4V1B2 Q9NB04 B4QML2 A0A1W4W6D1 A0A0M4EYE7 B3M7A0 A0A195F8V7 A0A1W4WZU9 B4IZX0 E9G0A2 N6TG89 U4UWR8 A0A1A9VPZ1 T1INI3 A0A1W4XMX9 A0A1B0BE50 J9LZD5 A0A1A9XN62 A0A1I8NX36 A0A182GL97 E9INJ5 A0A2A4K1J6 A0A3S2NZJ4 T1KYF5 A0A212ET84 A0A2A3E5I0 J9HI13 A0A2H1WVM7 A0A0T6ASQ8 A0A210QCE8 A0A2D5MEN1 A0A194PUS4 A0A2C9KCL4 A0A2C9KCJ1 A0A2C9KCM0 A0A2C9KCL7 A0A2S2Q8I5 A0A1B6E0P2 A0A1B6DSX3 A0A0K2SZ76 A0A2H5BFC6 A0A1E1XJ94 K1R7T1 A0A182QW64 R7V132 A0A2D5MCY4 A0A182N1G4 A0A1W0XEI4
A0A212ERH9 A0A0L7KSF9 E2B674 A0A0M8ZVG3 A0A158NCF0 F4WWZ8 A0A154PNZ1 A0A026X228 A0A195BWA7 A0A151JQU8 A0A1Y1M2Z1 A0A0L7R1E0 A0A2J7R5D1 A0A2J7R5D6 A0A2J7R5D2 A0A2J7R5D3 A0A1B6GQS6 A0A1B6HFI9 A0A1B6HTA6 A0A1B6ELN7 A0A087ZV81 D6WRN8 A0A151WMY2 A0A139WEE0 D6WPR0 A0A0P4VIU4 A0A146KZ93 A0A232FA68 A0A023F3W4 A0A224X4S0 A0A146LAK9 A0A151IBX1 E0VGE8 A0A3L8DGH0 A0A0A9YHT4 A0A182GIF1 A0A336KTG0 A0A1B0DAI8 A0A1D2MTJ7 B0WHT5 B4PDU5 A0A1Y1MNC1 A0A0J9RM08 D6W6M5 B3NEV8 A4V1B2 Q9NB04 B4QML2 A0A1W4W6D1 A0A0M4EYE7 B3M7A0 A0A195F8V7 A0A1W4WZU9 B4IZX0 E9G0A2 N6TG89 U4UWR8 A0A1A9VPZ1 T1INI3 A0A1W4XMX9 A0A1B0BE50 J9LZD5 A0A1A9XN62 A0A1I8NX36 A0A182GL97 E9INJ5 A0A2A4K1J6 A0A3S2NZJ4 T1KYF5 A0A212ET84 A0A2A3E5I0 J9HI13 A0A2H1WVM7 A0A0T6ASQ8 A0A210QCE8 A0A2D5MEN1 A0A194PUS4 A0A2C9KCL4 A0A2C9KCJ1 A0A2C9KCM0 A0A2C9KCL7 A0A2S2Q8I5 A0A1B6E0P2 A0A1B6DSX3 A0A0K2SZ76 A0A2H5BFC6 A0A1E1XJ94 K1R7T1 A0A182QW64 R7V132 A0A2D5MCY4 A0A182N1G4 A0A1W0XEI4
Pubmed
19121390
28756777
26354079
22118469
26227816
20798317
+ More
21347285 21719571 24508170 28004739 18362917 19820115 27129103 26823975 28648823 25474469 20566863 30249741 25401762 26483478 27289101 17994087 17550304 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 10102271 11076972 12537569 11740560 12900452 14667407 18327897 21292972 23537049 21282665 17510324 28812685 29337314 15562597 29228898 29209593 22992520 23254933
21347285 21719571 24508170 28004739 18362917 19820115 27129103 26823975 28648823 25474469 20566863 30249741 25401762 26483478 27289101 17994087 17550304 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 10102271 11076972 12537569 11740560 12900452 14667407 18327897 21292972 23537049 21282665 17510324 28812685 29337314 15562597 29228898 29209593 22992520 23254933
EMBL
BABH01034227
NWSH01001542
PCG70908.1
ODYU01000125
SOQ34374.1
KZ149945
+ More
PZC76817.1 KQ460205 KPJ17024.1 KQ459604 KPI92107.1 AGBW02013010 OWR44102.1 JTDY01006225 KOB66182.1 GL445930 EFN88767.1 KQ435830 KOX71767.1 ADTU01011857 ADTU01011858 ADTU01011859 ADTU01011860 ADTU01011861 ADTU01011862 ADTU01011863 ADTU01011864 ADTU01011865 ADTU01011866 GL888417 EGI61241.1 KQ435007 KZC13605.1 KK107034 EZA62056.1 KQ976396 KYM92914.1 KQ978619 KYN29768.1 GEZM01041885 GEZM01041880 JAV80119.1 KQ414668 KOC64659.1 NEVH01006994 PNF36039.1 PNF36040.1 PNF36041.1 PNF36042.1 GECZ01004998 JAS64771.1 GECU01035995 GECU01034273 JAS71711.1 JAS73433.1 GECU01029831 JAS77875.1 GECZ01030919 JAS38850.1 KQ971354 EFA07063.2 KQ982934 KYQ49168.1 KYB26282.1 EFA06857.2 GDKW01001773 JAI54822.1 GDHC01018599 JAQ00030.1 NNAY01000618 OXU27370.1 GBBI01002582 JAC16130.1 GFTR01008919 JAW07507.1 GDHC01014477 JAQ04152.1 KQ978076 KYM97225.1 DS235140 EEB12454.1 QOIP01000008 RLU19272.1 GBHO01012413 GBRD01008386 JAG31191.1 JAG57435.1 JXUM01066554 KQ562405 KXJ75988.1 UFQS01000466 UFQT01000466 SSX04240.1 SSX24605.1 AJVK01029121 LJIJ01000577 ODM96114.1 DS231939 EDS27952.1 CM000159 EDW92910.1 GEZM01027922 JAV86578.1 CM002912 KMY96953.1 KQ971307 EFA10862.2 CH954178 EDV50231.1 AE014296 AAF47573.1 AAG22224.1 AF103942 AF274350 AF132193 CM000363 EDX08869.1 CP012525 ALC43195.1 CH902618 EDV38761.1 KQ981727 KYN37040.1 CH916366 EDV96742.1 GL732528 EFX86868.1 APGK01039072 KB740967 ENN76793.1 KB632399 ERL94720.1 JH431176 JXJN01012795 ABLF02020507 JXUM01071603 JXUM01071604 JXUM01071605 KQ562674 KXJ75346.1 GL764306 EFZ17853.1 NWSH01000300 PCG77633.1 RSAL01000082 RVE48543.1 CAEY01000711 CAEY01000712 AGBW02012631 OWR44702.1 KZ288362 PBC26948.1 CH477277 EJY57494.1 ODYU01011401 SOQ57125.1 LJIG01022905 KRT78129.1 NEDP02004192 OWF46384.1 NYWD01000157 MAD20599.1 KQ459597 KPI94885.1 GGMS01004677 MBY73880.1 GEDC01005803 JAS31495.1 GEDC01008535 JAS28763.1 HACA01001326 CDW18687.1 MG197681 AUG84431.1 GFAA01004141 JAT99293.1 JH818426 EKC30021.1 AXCN02001376 AMQN01006189 KB297790 ELU09932.1 NYWD01000070 MAD20052.1 MTYJ01000001 OQV25884.1
PZC76817.1 KQ460205 KPJ17024.1 KQ459604 KPI92107.1 AGBW02013010 OWR44102.1 JTDY01006225 KOB66182.1 GL445930 EFN88767.1 KQ435830 KOX71767.1 ADTU01011857 ADTU01011858 ADTU01011859 ADTU01011860 ADTU01011861 ADTU01011862 ADTU01011863 ADTU01011864 ADTU01011865 ADTU01011866 GL888417 EGI61241.1 KQ435007 KZC13605.1 KK107034 EZA62056.1 KQ976396 KYM92914.1 KQ978619 KYN29768.1 GEZM01041885 GEZM01041880 JAV80119.1 KQ414668 KOC64659.1 NEVH01006994 PNF36039.1 PNF36040.1 PNF36041.1 PNF36042.1 GECZ01004998 JAS64771.1 GECU01035995 GECU01034273 JAS71711.1 JAS73433.1 GECU01029831 JAS77875.1 GECZ01030919 JAS38850.1 KQ971354 EFA07063.2 KQ982934 KYQ49168.1 KYB26282.1 EFA06857.2 GDKW01001773 JAI54822.1 GDHC01018599 JAQ00030.1 NNAY01000618 OXU27370.1 GBBI01002582 JAC16130.1 GFTR01008919 JAW07507.1 GDHC01014477 JAQ04152.1 KQ978076 KYM97225.1 DS235140 EEB12454.1 QOIP01000008 RLU19272.1 GBHO01012413 GBRD01008386 JAG31191.1 JAG57435.1 JXUM01066554 KQ562405 KXJ75988.1 UFQS01000466 UFQT01000466 SSX04240.1 SSX24605.1 AJVK01029121 LJIJ01000577 ODM96114.1 DS231939 EDS27952.1 CM000159 EDW92910.1 GEZM01027922 JAV86578.1 CM002912 KMY96953.1 KQ971307 EFA10862.2 CH954178 EDV50231.1 AE014296 AAF47573.1 AAG22224.1 AF103942 AF274350 AF132193 CM000363 EDX08869.1 CP012525 ALC43195.1 CH902618 EDV38761.1 KQ981727 KYN37040.1 CH916366 EDV96742.1 GL732528 EFX86868.1 APGK01039072 KB740967 ENN76793.1 KB632399 ERL94720.1 JH431176 JXJN01012795 ABLF02020507 JXUM01071603 JXUM01071604 JXUM01071605 KQ562674 KXJ75346.1 GL764306 EFZ17853.1 NWSH01000300 PCG77633.1 RSAL01000082 RVE48543.1 CAEY01000711 CAEY01000712 AGBW02012631 OWR44702.1 KZ288362 PBC26948.1 CH477277 EJY57494.1 ODYU01011401 SOQ57125.1 LJIG01022905 KRT78129.1 NEDP02004192 OWF46384.1 NYWD01000157 MAD20599.1 KQ459597 KPI94885.1 GGMS01004677 MBY73880.1 GEDC01005803 JAS31495.1 GEDC01008535 JAS28763.1 HACA01001326 CDW18687.1 MG197681 AUG84431.1 GFAA01004141 JAT99293.1 JH818426 EKC30021.1 AXCN02001376 AMQN01006189 KB297790 ELU09932.1 NYWD01000070 MAD20052.1 MTYJ01000001 OQV25884.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000007151
UP000037510
+ More
UP000008237 UP000053105 UP000005205 UP000007755 UP000076502 UP000053097 UP000078540 UP000078492 UP000053825 UP000235965 UP000005203 UP000007266 UP000075809 UP000215335 UP000078542 UP000009046 UP000279307 UP000069940 UP000249989 UP000092462 UP000094527 UP000002320 UP000002282 UP000008711 UP000000803 UP000000304 UP000192221 UP000092553 UP000007801 UP000078541 UP000192223 UP000001070 UP000000305 UP000019118 UP000030742 UP000078200 UP000092460 UP000007819 UP000092443 UP000095300 UP000283053 UP000015104 UP000242457 UP000008820 UP000242188 UP000076420 UP000005408 UP000075886 UP000014760 UP000075884
UP000008237 UP000053105 UP000005205 UP000007755 UP000076502 UP000053097 UP000078540 UP000078492 UP000053825 UP000235965 UP000005203 UP000007266 UP000075809 UP000215335 UP000078542 UP000009046 UP000279307 UP000069940 UP000249989 UP000092462 UP000094527 UP000002320 UP000002282 UP000008711 UP000000803 UP000000304 UP000192221 UP000092553 UP000007801 UP000078541 UP000192223 UP000001070 UP000000305 UP000019118 UP000030742 UP000078200 UP000092460 UP000007819 UP000092443 UP000095300 UP000283053 UP000015104 UP000242457 UP000008820 UP000242188 UP000076420 UP000005408 UP000075886 UP000014760 UP000075884
Interpro
ProteinModelPortal
H9IU82
A0A2A4JGR2
A0A2H1V279
A0A2W1BUE1
A0A194RHV9
A0A194PFP2
+ More
A0A212ERH9 A0A0L7KSF9 E2B674 A0A0M8ZVG3 A0A158NCF0 F4WWZ8 A0A154PNZ1 A0A026X228 A0A195BWA7 A0A151JQU8 A0A1Y1M2Z1 A0A0L7R1E0 A0A2J7R5D1 A0A2J7R5D6 A0A2J7R5D2 A0A2J7R5D3 A0A1B6GQS6 A0A1B6HFI9 A0A1B6HTA6 A0A1B6ELN7 A0A087ZV81 D6WRN8 A0A151WMY2 A0A139WEE0 D6WPR0 A0A0P4VIU4 A0A146KZ93 A0A232FA68 A0A023F3W4 A0A224X4S0 A0A146LAK9 A0A151IBX1 E0VGE8 A0A3L8DGH0 A0A0A9YHT4 A0A182GIF1 A0A336KTG0 A0A1B0DAI8 A0A1D2MTJ7 B0WHT5 B4PDU5 A0A1Y1MNC1 A0A0J9RM08 D6W6M5 B3NEV8 A4V1B2 Q9NB04 B4QML2 A0A1W4W6D1 A0A0M4EYE7 B3M7A0 A0A195F8V7 A0A1W4WZU9 B4IZX0 E9G0A2 N6TG89 U4UWR8 A0A1A9VPZ1 T1INI3 A0A1W4XMX9 A0A1B0BE50 J9LZD5 A0A1A9XN62 A0A1I8NX36 A0A182GL97 E9INJ5 A0A2A4K1J6 A0A3S2NZJ4 T1KYF5 A0A212ET84 A0A2A3E5I0 J9HI13 A0A2H1WVM7 A0A0T6ASQ8 A0A210QCE8 A0A2D5MEN1 A0A194PUS4 A0A2C9KCL4 A0A2C9KCJ1 A0A2C9KCM0 A0A2C9KCL7 A0A2S2Q8I5 A0A1B6E0P2 A0A1B6DSX3 A0A0K2SZ76 A0A2H5BFC6 A0A1E1XJ94 K1R7T1 A0A182QW64 R7V132 A0A2D5MCY4 A0A182N1G4 A0A1W0XEI4
A0A212ERH9 A0A0L7KSF9 E2B674 A0A0M8ZVG3 A0A158NCF0 F4WWZ8 A0A154PNZ1 A0A026X228 A0A195BWA7 A0A151JQU8 A0A1Y1M2Z1 A0A0L7R1E0 A0A2J7R5D1 A0A2J7R5D6 A0A2J7R5D2 A0A2J7R5D3 A0A1B6GQS6 A0A1B6HFI9 A0A1B6HTA6 A0A1B6ELN7 A0A087ZV81 D6WRN8 A0A151WMY2 A0A139WEE0 D6WPR0 A0A0P4VIU4 A0A146KZ93 A0A232FA68 A0A023F3W4 A0A224X4S0 A0A146LAK9 A0A151IBX1 E0VGE8 A0A3L8DGH0 A0A0A9YHT4 A0A182GIF1 A0A336KTG0 A0A1B0DAI8 A0A1D2MTJ7 B0WHT5 B4PDU5 A0A1Y1MNC1 A0A0J9RM08 D6W6M5 B3NEV8 A4V1B2 Q9NB04 B4QML2 A0A1W4W6D1 A0A0M4EYE7 B3M7A0 A0A195F8V7 A0A1W4WZU9 B4IZX0 E9G0A2 N6TG89 U4UWR8 A0A1A9VPZ1 T1INI3 A0A1W4XMX9 A0A1B0BE50 J9LZD5 A0A1A9XN62 A0A1I8NX36 A0A182GL97 E9INJ5 A0A2A4K1J6 A0A3S2NZJ4 T1KYF5 A0A212ET84 A0A2A3E5I0 J9HI13 A0A2H1WVM7 A0A0T6ASQ8 A0A210QCE8 A0A2D5MEN1 A0A194PUS4 A0A2C9KCL4 A0A2C9KCJ1 A0A2C9KCM0 A0A2C9KCL7 A0A2S2Q8I5 A0A1B6E0P2 A0A1B6DSX3 A0A0K2SZ76 A0A2H5BFC6 A0A1E1XJ94 K1R7T1 A0A182QW64 R7V132 A0A2D5MCY4 A0A182N1G4 A0A1W0XEI4
PDB
2DLU
E-value=3.01071e-24,
Score=279
Ontologies
GO
GO:0016327
GO:0032033
GO:0035088
GO:0008594
GO:0045494
GO:0035509
GO:0016334
GO:0005912
GO:0034334
GO:0035003
GO:0001736
GO:0016324
GO:0005080
GO:0035209
GO:0005886
GO:0007043
GO:0045176
GO:0034332
GO:0016333
GO:0002009
GO:0005737
GO:0045179
GO:0005918
GO:0007163
GO:0005635
GO:0016332
GO:0005515
GO:0006355
GO:0043565
GO:0006886
GO:0016192
GO:0030117
GO:0007154
GO:0016020
GO:0005112
GO:0015930
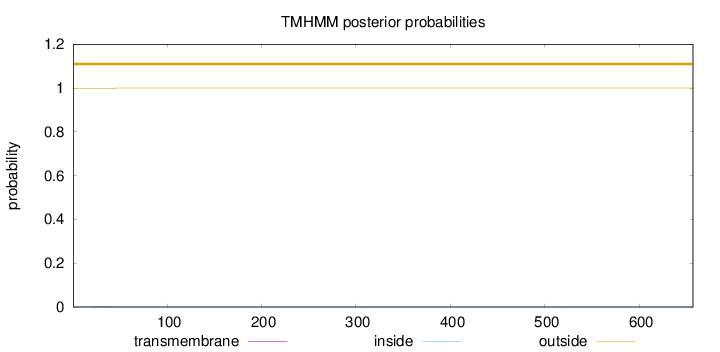
Topology
Subcellular location
Membrane
Length:
657
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.03684
Exp number, first 60 AAs:
0.0368
Total prob of N-in:
0.00197
outside
1 - 657
Population Genetic Test Statistics
Pi
13.274902
Theta
12.813541
Tajima's D
-0.715623
CLR
2.31603
CSRT
0.1998400079996
Interpretation
Uncertain
