Gene
KWMTBOMO14948
Pre Gene Modal
BGIBMGA000762
Annotation
PREDICTED:_thyroid_transcription_factor_1-like_isoform_X1_[Bombyx_mori]
Transcription factor
Location in the cell
Nuclear Reliability : 4.082
Sequence
CDS
ATGGGCGGGCTGGCGGCGTGTGCAGGTCCTGACGCCAAGCCCATGCAATTCCCGTTGGCGCAGCGGCGGAAGAGGAGAGTGCTCTTTACACAAGCCCAGGTTTACGAGCTGGAGCGGAGATTTAAGCAGCAGAAGTATCTGTCGGCGCCGGAGAGGGAGCACCTCGCTTCGCTCATTCACCTAACGCCGACCCAAGTCAAAATATGGTTTCAAAACCATCGTTACAAATGTAAGAGACAAGCTAAAGAGAAAGCAATGGCGGAACAGAACCAACATAATCAGGTAACGTATTGCCCGCAGTCGTCATCGTCGCCGCGACGGGTCGCAGTTCCCGTGCTGGTGAAGGACGGCAAGCCGTGCGGGGCGGGCGAGTCGTCCTCGCCGGCGCACAGCCACTGCGCCACGCCCACCTCCGGGTACTCGGCCCCTCAGGGCTCGCAGGCCGCCTGCAGCAACCCGCTGGTCTCCTCGTACCGGCAGCCCGCCGCGTACCAGCAGCAATGCTCGGGATACCTGCCGCTACAGGGTCGAGCCTGGTAG
Protein
MGGLAACAGPDAKPMQFPLAQRRKRRVLFTQAQVYELERRFKQQKYLSAPEREHLASLIHLTPTQVKIWFQNHRYKCKRQAKEKAMAEQNQHNQVTYCPQSSSSPRRVAVPVLVKDGKPCGAGESSSPAHSHCATPTSGYSAPQGSQAACSNPLVSSYRQPAAYQQQCSGYLPLQGRAW
Summary
Uniprot
A0A2H1WDM0
A0A2A4K7T4
A0A2W1BT56
A0A194PGF3
A0A212EYG8
A0A1B0CEF2
+ More
A0A1W4VYH7 A0A0R1E856 B4IV98 A0A0P9A989 A0A336N2U9 B3NJ53 A8Y527 B4GMA5 A0A3B0KT07 Q7PL95 A0A1J1IRY9 B3MWX3 A0A1A9W3K8 Q9NCG1 A0A336LTW9 D6WPZ8 B4KAU6 W8BN78 B4JHS5 B4NA09 B4LVR0 A0A1A9XZC3 A0A1B0BJE1 A0A1A9VEP0 A0A1A9ZXJ9 A0A0R1E8Q8 A0A0Q9WDS9 W8C9W6 A0A1Y9HAJ6 A0A034WVQ1 A0A1B0FAA0 A0A0K8W1D0 A0A0K8U9P1 A0A2J7PKS8 A0A0A1X2E8 A0A182LTG7 A0A026VVB2 T1JDL6 A0A195BNG0 A0A158NKN6 E0VIY7 A0A195CKW2 A0A2S2Q9G8 A0A2H8TGP7 J9JLF2 A0A0C9Q4P7 A0A2S2P6H3 A0A2A3ERV4 K7JAH5 A0A088A0E0 A0A3P9A4B5 A0A1S3S7P2 A0A3P8YF76 Q58A08 A0A3B3ZH48 A0A1S3M983 A0A226P406 Q58A09 A0A3Q3FAV0 A0A2D0Q012 A0A226MU60 A0A3B3S1H1 A0A3B3TAQ6 A0A3Q2X202
A0A1W4VYH7 A0A0R1E856 B4IV98 A0A0P9A989 A0A336N2U9 B3NJ53 A8Y527 B4GMA5 A0A3B0KT07 Q7PL95 A0A1J1IRY9 B3MWX3 A0A1A9W3K8 Q9NCG1 A0A336LTW9 D6WPZ8 B4KAU6 W8BN78 B4JHS5 B4NA09 B4LVR0 A0A1A9XZC3 A0A1B0BJE1 A0A1A9VEP0 A0A1A9ZXJ9 A0A0R1E8Q8 A0A0Q9WDS9 W8C9W6 A0A1Y9HAJ6 A0A034WVQ1 A0A1B0FAA0 A0A0K8W1D0 A0A0K8U9P1 A0A2J7PKS8 A0A0A1X2E8 A0A182LTG7 A0A026VVB2 T1JDL6 A0A195BNG0 A0A158NKN6 E0VIY7 A0A195CKW2 A0A2S2Q9G8 A0A2H8TGP7 J9JLF2 A0A0C9Q4P7 A0A2S2P6H3 A0A2A3ERV4 K7JAH5 A0A088A0E0 A0A3P9A4B5 A0A1S3S7P2 A0A3P8YF76 Q58A08 A0A3B3ZH48 A0A1S3M983 A0A226P406 Q58A09 A0A3Q3FAV0 A0A2D0Q012 A0A226MU60 A0A3B3S1H1 A0A3B3TAQ6 A0A3Q2X202
Pubmed
EMBL
ODYU01007941
SOQ51151.1
NWSH01000070
PCG79974.1
KZ149945
PZC76815.1
+ More
KQ459604 KPI92108.1 AGBW02011506 OWR46543.1 AJWK01008827 AJWK01008828 AJWK01008829 CH892673 KRK05528.1 EDX00263.2 CH902627 KPU74873.1 UFQS01002127 UFQT01002127 SSX13282.1 SSX32718.1 CH954178 EDV52836.2 AE014296 EDP28066.2 CH479185 EDW37979.1 OUUW01000014 SPP88361.1 EAA45970.1 CVRI01000059 CRL03005.1 EDV33953.2 KPU74875.1 AF220096 AAF26436.1 UFQT01000189 SSX21375.1 KQ971354 EFA07447.2 CH933806 EDW13619.1 KRG00508.1 KRG00509.1 GAMC01006293 GAMC01006289 JAC00263.1 CH916369 EDV93914.1 CH964232 EDW81764.2 CH940650 EDW66483.1 JXJN01015373 JXJN01015374 KRK05529.1 KRF82707.1 GAMC01006295 GAMC01006292 GAMC01006291 JAC00265.1 AXCN02001164 GAKP01001089 JAC57863.1 CCAG010015604 GDHF01007420 JAI44894.1 GDHF01029016 GDHF01016634 GDHF01008293 GDHF01004783 GDHF01000477 JAI23298.1 JAI35680.1 JAI44021.1 JAI47531.1 JAI51837.1 NEVH01024531 PNF16935.1 GBXI01009226 JAD05066.1 AXCM01004066 AXCM01004067 KK107796 EZA47723.1 JH432107 KQ976439 KYM86736.1 ADTU01018930 ADTU01018931 ADTU01018932 ADTU01018933 ADTU01018934 ADTU01018935 DS235215 EEB13343.1 KQ977649 KYN00719.1 GGMS01005164 MBY74367.1 GFXV01001406 MBW13211.1 ABLF02019158 GBYB01009058 JAG78825.1 GGMR01012396 MBY25015.1 KZ288194 PBC33946.1 AAZX01004917 AAZX01008692 AB159044 BAD93686.1 AWGT02000182 OXB74320.1 AB159043 BAD93685.1 MCFN01000463 OXB58539.1
KQ459604 KPI92108.1 AGBW02011506 OWR46543.1 AJWK01008827 AJWK01008828 AJWK01008829 CH892673 KRK05528.1 EDX00263.2 CH902627 KPU74873.1 UFQS01002127 UFQT01002127 SSX13282.1 SSX32718.1 CH954178 EDV52836.2 AE014296 EDP28066.2 CH479185 EDW37979.1 OUUW01000014 SPP88361.1 EAA45970.1 CVRI01000059 CRL03005.1 EDV33953.2 KPU74875.1 AF220096 AAF26436.1 UFQT01000189 SSX21375.1 KQ971354 EFA07447.2 CH933806 EDW13619.1 KRG00508.1 KRG00509.1 GAMC01006293 GAMC01006289 JAC00263.1 CH916369 EDV93914.1 CH964232 EDW81764.2 CH940650 EDW66483.1 JXJN01015373 JXJN01015374 KRK05529.1 KRF82707.1 GAMC01006295 GAMC01006292 GAMC01006291 JAC00265.1 AXCN02001164 GAKP01001089 JAC57863.1 CCAG010015604 GDHF01007420 JAI44894.1 GDHF01029016 GDHF01016634 GDHF01008293 GDHF01004783 GDHF01000477 JAI23298.1 JAI35680.1 JAI44021.1 JAI47531.1 JAI51837.1 NEVH01024531 PNF16935.1 GBXI01009226 JAD05066.1 AXCM01004066 AXCM01004067 KK107796 EZA47723.1 JH432107 KQ976439 KYM86736.1 ADTU01018930 ADTU01018931 ADTU01018932 ADTU01018933 ADTU01018934 ADTU01018935 DS235215 EEB13343.1 KQ977649 KYN00719.1 GGMS01005164 MBY74367.1 GFXV01001406 MBW13211.1 ABLF02019158 GBYB01009058 JAG78825.1 GGMR01012396 MBY25015.1 KZ288194 PBC33946.1 AAZX01004917 AAZX01008692 AB159044 BAD93686.1 AWGT02000182 OXB74320.1 AB159043 BAD93685.1 MCFN01000463 OXB58539.1
Proteomes
UP000218220
UP000053268
UP000007151
UP000092461
UP000192221
UP000002282
+ More
UP000007801 UP000008711 UP000000803 UP000008744 UP000268350 UP000183832 UP000091820 UP000007266 UP000009192 UP000001070 UP000007798 UP000008792 UP000092443 UP000092460 UP000078200 UP000092445 UP000075886 UP000092444 UP000235965 UP000075883 UP000053097 UP000078540 UP000005205 UP000009046 UP000078542 UP000007819 UP000242457 UP000002358 UP000005203 UP000265140 UP000087266 UP000261520 UP000198419 UP000261660 UP000221080 UP000198323 UP000261540 UP000264840
UP000007801 UP000008711 UP000000803 UP000008744 UP000268350 UP000183832 UP000091820 UP000007266 UP000009192 UP000001070 UP000007798 UP000008792 UP000092443 UP000092460 UP000078200 UP000092445 UP000075886 UP000092444 UP000235965 UP000075883 UP000053097 UP000078540 UP000005205 UP000009046 UP000078542 UP000007819 UP000242457 UP000002358 UP000005203 UP000265140 UP000087266 UP000261520 UP000198419 UP000261660 UP000221080 UP000198323 UP000261540 UP000264840
Pfam
PF00046 Homeodomain
Interpro
SUPFAM
SSF46689
SSF46689
ProteinModelPortal
A0A2H1WDM0
A0A2A4K7T4
A0A2W1BT56
A0A194PGF3
A0A212EYG8
A0A1B0CEF2
+ More
A0A1W4VYH7 A0A0R1E856 B4IV98 A0A0P9A989 A0A336N2U9 B3NJ53 A8Y527 B4GMA5 A0A3B0KT07 Q7PL95 A0A1J1IRY9 B3MWX3 A0A1A9W3K8 Q9NCG1 A0A336LTW9 D6WPZ8 B4KAU6 W8BN78 B4JHS5 B4NA09 B4LVR0 A0A1A9XZC3 A0A1B0BJE1 A0A1A9VEP0 A0A1A9ZXJ9 A0A0R1E8Q8 A0A0Q9WDS9 W8C9W6 A0A1Y9HAJ6 A0A034WVQ1 A0A1B0FAA0 A0A0K8W1D0 A0A0K8U9P1 A0A2J7PKS8 A0A0A1X2E8 A0A182LTG7 A0A026VVB2 T1JDL6 A0A195BNG0 A0A158NKN6 E0VIY7 A0A195CKW2 A0A2S2Q9G8 A0A2H8TGP7 J9JLF2 A0A0C9Q4P7 A0A2S2P6H3 A0A2A3ERV4 K7JAH5 A0A088A0E0 A0A3P9A4B5 A0A1S3S7P2 A0A3P8YF76 Q58A08 A0A3B3ZH48 A0A1S3M983 A0A226P406 Q58A09 A0A3Q3FAV0 A0A2D0Q012 A0A226MU60 A0A3B3S1H1 A0A3B3TAQ6 A0A3Q2X202
A0A1W4VYH7 A0A0R1E856 B4IV98 A0A0P9A989 A0A336N2U9 B3NJ53 A8Y527 B4GMA5 A0A3B0KT07 Q7PL95 A0A1J1IRY9 B3MWX3 A0A1A9W3K8 Q9NCG1 A0A336LTW9 D6WPZ8 B4KAU6 W8BN78 B4JHS5 B4NA09 B4LVR0 A0A1A9XZC3 A0A1B0BJE1 A0A1A9VEP0 A0A1A9ZXJ9 A0A0R1E8Q8 A0A0Q9WDS9 W8C9W6 A0A1Y9HAJ6 A0A034WVQ1 A0A1B0FAA0 A0A0K8W1D0 A0A0K8U9P1 A0A2J7PKS8 A0A0A1X2E8 A0A182LTG7 A0A026VVB2 T1JDL6 A0A195BNG0 A0A158NKN6 E0VIY7 A0A195CKW2 A0A2S2Q9G8 A0A2H8TGP7 J9JLF2 A0A0C9Q4P7 A0A2S2P6H3 A0A2A3ERV4 K7JAH5 A0A088A0E0 A0A3P9A4B5 A0A1S3S7P2 A0A3P8YF76 Q58A08 A0A3B3ZH48 A0A1S3M983 A0A226P406 Q58A09 A0A3Q3FAV0 A0A2D0Q012 A0A226MU60 A0A3B3S1H1 A0A3B3TAQ6 A0A3Q2X202
PDB
1FTT
E-value=5.37326e-23,
Score=262
Ontologies
GO
Topology
Subcellular location
Nucleus
Length:
179
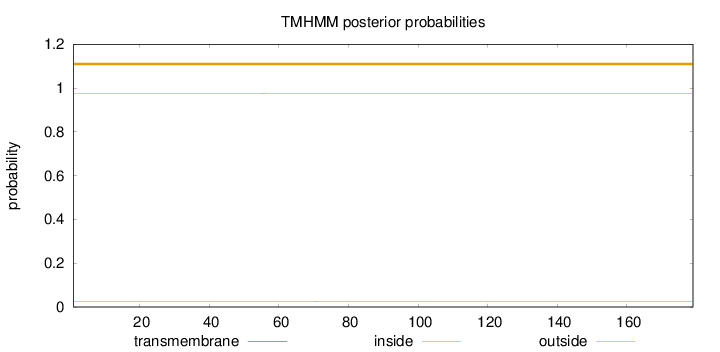
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00181
Exp number, first 60 AAs:
0.00069
Total prob of N-in:
0.02564
outside
1 - 179
Population Genetic Test Statistics
Pi
173.449962
Theta
168.396863
Tajima's D
0.270832
CLR
1.030159
CSRT
0.44322783860807
Interpretation
Possibly Positive selection
