Gene
KWMTBOMO14944
Pre Gene Modal
BGIBMGA000765
Annotation
PREDICTED:_ribosome-releasing_factor_2?_mitochondrial_[Amyelois_transitella]
Full name
Ribosome-releasing factor 2, mitochondrial
Alternative Name
Elongation factor G 2, mitochondrial
Elongation factor G2
Elongation factor G2
Location in the cell
Cytoplasmic Reliability : 2.211
Sequence
CDS
ATGGAAAATATTCGTAATATTGGTATTTTAGCGCACATTGATGCAGGTAAAACAACGACAACAGAAAGAATGCTGTTTTACTCAGGCACCATCAGATCTATGGGAGAGGTGCATCATGGGAATACCGTCACAGATTACATGGAACAAGAGAGGCAAAGGGGAATCACAATCACTTCTGCAGCAGTAACAATCCCTTGGAGAGGAGGGCAGATCAATCTAATAGATACACCAGGACACATAGACTTCACAATGGAGGTCGAACAGAGTTTGGCTGTTCTCGATGGTGCAGTTATAGTCCTTGATGGATCTGCCGGTGTCGAAGCACAAACATTAACAGTGTGGAGACAGGCCATCGGGTACAGAGTACCTCGGATACTCTACTTAAACAAGATGGACAGGAATGACGCATTCGTGGAGGCTTGCGTGAACTCGGTCACAGAGAAGCTGCAAGCCACGCCTTTGCTACTCCACCACACCGTTAGGCACGAAGGACGACTCATTGGACTAATAGATTTAATTAACCTAGAAGAGATAATATGGACTCAAGGACGAGGACAGAAGTTCACTCGAAGAAAACTGACGGAGAAAGACGACGGTCACAAATGGGAGGCGGCCGTTACTGACCATAGACAACTCGTGGACACTCTCTCTTCTATAGACGATGAGATAGCAGAGACAATCATAAACAACGAGTCTTTGGAGTTGAGCGCACGCGACATAGACAATGCGGTCAGAAGATCTACTATTAAAATGAAAGCTTTTCCCATTTTGTGTGGAAGTTCTTATAAAAATATAGGGGTTCAAACCTTGATGGACGGTGTCATGTCATATCTTCCAAGTCCCCTGGAAGGACACGAGCTGTATAAATGTTTCGGTGAGGAGCTAGCGGGCAGGGCTTTCAAAGTGATCCACGACGACCAGAGGGGAGTGCTGACCTTCGTGAGGCTGTACAGCGGCGAGATGAAGAAGGCCCAGAAGATCTACAACCTTGGACAGGACAGAAGCGAGCAGACGGGCGCGCTGTACGTGGCGTTGGCTGACGAGTATCGTCCAGTGGAGTCAGTGGCCGCGGGCAACATCGCCGTCGTGGGGTCACTCAAGGCGACCATGACGGGGGACCTGGTCACGTCCACACAAGCGGCGGCCAACAGGGCCCGGGGTCGGCTGGCGACGCTGCTCCGCCTCCCCGGGCGGGCGGAGGAGCTGATGCTGCCCAGCGCCAGGCAGAGGCTGCAGGCTCTCGACTCTGAACCAGAGCCTGAGGAACTTGCAGAAATTCTGCTCGGCATCGGCACCACCGTGCCCGAGCCCGTGTTCCTGTGCTCCATCGAGCCCCCCAGCGCCATGCACCAGGCGGCGCTGGAGACGGCGCTGGAGCAGCTGCAGCGGGAGGACCCGAGCCTCAGGGTCAACGCGGATGACGAAAGCGGACAGATCGTATTGGCCGGTATGGGCGAGCTCCACCTGGAAATCATAAAGGAGCGCATCATCAGAGAGTACAAGATAGACGTGGAGCTGGGCCCGCTGCAGATAGCGTACCGGGAGGCGCTGGTCTCTTCCGGCAAGAACACGCTCACTGTGGACAGGAAGATCGGCGGCGCCAGGCAGCAGCTGAAGGTCACCATGTCCGCCAGGACCGTCAAGGGAGTCGCTCAGGATAAGATCCTCAGGTTAGATAAGACGGTCGAGAGCGCGTCGAACCTCGCGCATCTGCACCCGCGGCAGCTGCAGGCCGTGCGGCAGGGGGTCGCCGCCGCGCTGCTGCACGGGCCCAAGCTCGGCTGTCCGGTGGTGGACGTGCAGGTGACGCTGCACTGGTTCGAGTCCGGGCGCGGCACCTCCGACTCCGTAGTGACGGCGTCCGTCGCCCAGTGCTTGAGGAAGGTGTTCGAAGAGGCGGACTCGATCCTCCTGGAGCCGGTGATGTCGCTGGAGGTGGTGTGTCCCGAGACGCACTCTCAGAGGGTGCTGGCCGACCTCTCGCGGCGGAGGGTCGAGGTGCAGCACATCCAGCTGCGACAACACAACAAGGTGATAGAGTGCATAGCTCCGCTGTCGGAGGTGGTGGGCTACTCGTCGACGCTGCGCTCGCTGAGCTCGGGGCTGGCCACCTTCTCCATGCAGTTCCACTCGCACCGCCAGATGGCGCCGCAACACGAGCAGCTCGCCGTCAAGAATGTCACCGGGTTCTAG
Protein
MENIRNIGILAHIDAGKTTTTERMLFYSGTIRSMGEVHHGNTVTDYMEQERQRGITITSAAVTIPWRGGQINLIDTPGHIDFTMEVEQSLAVLDGAVIVLDGSAGVEAQTLTVWRQAIGYRVPRILYLNKMDRNDAFVEACVNSVTEKLQATPLLLHHTVRHEGRLIGLIDLINLEEIIWTQGRGQKFTRRKLTEKDDGHKWEAAVTDHRQLVDTLSSIDDEIAETIINNESLELSARDIDNAVRRSTIKMKAFPILCGSSYKNIGVQTLMDGVMSYLPSPLEGHELYKCFGEELAGRAFKVIHDDQRGVLTFVRLYSGEMKKAQKIYNLGQDRSEQTGALYVALADEYRPVESVAAGNIAVVGSLKATMTGDLVTSTQAAANRARGRLATLLRLPGRAEELMLPSARQRLQALDSEPEPEELAEILLGIGTTVPEPVFLCSIEPPSAMHQAALETALEQLQREDPSLRVNADDESGQIVLAGMGELHLEIIKERIIREYKIDVELGPLQIAYREALVSSGKNTLTVDRKIGGARQQLKVTMSARTVKGVAQDKILRLDKTVESASNLAHLHPRQLQAVRQGVAAALLHGPKLGCPVVDVQVTLHWFESGRGTSDSVVTASVAQCLRKVFEEADSILLEPVMSLEVVCPETHSQRVLADLSRRRVEVQHIQLRQHNKVIECIAPLSEVVGYSSTLRSLSSGLATFSMQFHSHRQMAPQHEQLAVKNVTGF
Summary
Description
Mitochondrial GTPase that mediates the disassembly of ribosomes from messenger RNA at the termination of mitochondrial protein biosynthesis. Not involved in the GTP-dependent ribosomal translocation step during translation elongation.
Mitochondrial GTPase that mediates the disassembly of ribosomes from messenger RNA at the termination of mitochondrial protein biosynthesis. Acts in collaboration with MRRF. GTP hydrolysis follows the ribosome disassembly and probably occurs on the ribosome large subunit. Not involved in the GTP-dependent ribosomal translocation step during translation elongation.
Mitochondrial GTPase that mediates the disassembly of ribosomes from messenger RNA at the termination of mitochondrial protein biosynthesis. Acts in collaboration with MRRF. GTP hydrolysis follows the ribosome disassembly and probably occurs on the ribosome large subunit. Not involved in the GTP-dependent ribosomal translocation step during translation elongation.
Similarity
Belongs to the TRAFAC class translation factor GTPase superfamily. Classic translation factor GTPase family. EF-G/EF-2 subfamily.
Keywords
Complete proteome
GTP-binding
Mitochondrion
Nucleotide-binding
Protein biosynthesis
Reference proteome
Transit peptide
Feature
chain Ribosome-releasing factor 2, mitochondrial
Uniprot
H9IU35
A0A2W1BYG6
A0A2A4J5K6
A0A194PFP7
A0A2H1WFC6
A0A3S2M9K1
+ More
A0A212EYL2 A0A194RGT7 A0A1B6CPE1 A0A2P8Y271 A0A151WQ12 A0A195C310 A0A158NJ41 A0A195DJU9 F4W794 A0A195BL22 A0A0C9REE6 E1ZVL4 D6WPX8 A0A1B6J8A0 E2BK05 A0A0L7KHK7 A0A1B6GLX8 A0A026VVW9 A0A1Y1N3N6 A0A3L8DDX8 E9IT92 A0A1B6KGD5 J9JKG8 A0A232FGQ4 A0A087ZT96 A0A067REY6 A0A195EYS5 K7J309 A0A3R7QK99 A0A1B6EU19 A0A0M9A4I1 N6UEF1 A0A0Q9XCL1 A0A0J7L6I1 B4JSI3 B4NAU8 A0A0P4W9E1 Q29BD5 B4GNT0 B3M011 A0A0M3QXL7 A0A1A9VVW3 A0A3B0JJ22 A0A1A9Z3R1 A0A1A9XFK6 A0A1J1J0F0 A0A0L7QMD6 A0A1Y1MZ82 A0A1W4W1J0 A0A1A9WRW7 A0A0K8UAG6 B3P8M3 A0A0A1X4W8 Q9VCX4 A0A0L0BS17 A0A1I8MHD4 B0W010 B4M416 I1WYH6 A0A226F0P9 A0A0P6IYK0 A0A1S4FZT8 A0A1Q3F921 A0A1I8PK89 A0A182TL00 A0A182WTC1 A0A182HVB4 A0A182M335 A0A182L264 A0A182VKM0 A0A1S4GWL9 A0A293LH03 A0A182GKZ0 A0A182GL48 A0A0P5HXK6 A0A0P6BC28 A0A182W9S8 A0A0P6IQC6 A0A182ST14 A0A182NE67 A0A1D2MVA9 A0A3S1BRK1 A0A182QEH8 A0A164LWQ0 A0A182RBQ2 E9FU13 A0A182YGX6 A0A2A3EC44 F7G7T5 T1HV44 A0A0K8UG02 A0A0N8CEK7 A0A154P443 C3YQ46
A0A212EYL2 A0A194RGT7 A0A1B6CPE1 A0A2P8Y271 A0A151WQ12 A0A195C310 A0A158NJ41 A0A195DJU9 F4W794 A0A195BL22 A0A0C9REE6 E1ZVL4 D6WPX8 A0A1B6J8A0 E2BK05 A0A0L7KHK7 A0A1B6GLX8 A0A026VVW9 A0A1Y1N3N6 A0A3L8DDX8 E9IT92 A0A1B6KGD5 J9JKG8 A0A232FGQ4 A0A087ZT96 A0A067REY6 A0A195EYS5 K7J309 A0A3R7QK99 A0A1B6EU19 A0A0M9A4I1 N6UEF1 A0A0Q9XCL1 A0A0J7L6I1 B4JSI3 B4NAU8 A0A0P4W9E1 Q29BD5 B4GNT0 B3M011 A0A0M3QXL7 A0A1A9VVW3 A0A3B0JJ22 A0A1A9Z3R1 A0A1A9XFK6 A0A1J1J0F0 A0A0L7QMD6 A0A1Y1MZ82 A0A1W4W1J0 A0A1A9WRW7 A0A0K8UAG6 B3P8M3 A0A0A1X4W8 Q9VCX4 A0A0L0BS17 A0A1I8MHD4 B0W010 B4M416 I1WYH6 A0A226F0P9 A0A0P6IYK0 A0A1S4FZT8 A0A1Q3F921 A0A1I8PK89 A0A182TL00 A0A182WTC1 A0A182HVB4 A0A182M335 A0A182L264 A0A182VKM0 A0A1S4GWL9 A0A293LH03 A0A182GKZ0 A0A182GL48 A0A0P5HXK6 A0A0P6BC28 A0A182W9S8 A0A0P6IQC6 A0A182ST14 A0A182NE67 A0A1D2MVA9 A0A3S1BRK1 A0A182QEH8 A0A164LWQ0 A0A182RBQ2 E9FU13 A0A182YGX6 A0A2A3EC44 F7G7T5 T1HV44 A0A0K8UG02 A0A0N8CEK7 A0A154P443 C3YQ46
Pubmed
19121390
28756777
26354079
22118469
29403074
21347285
+ More
21719571 20798317 18362917 19820115 26227816 24508170 28004739 30249741 21282665 28648823 24845553 20075255 23537049 17994087 15632085 25830018 20132446 21364917 10731132 12537572 26108605 25315136 26999592 17510324 20966253 12364791 26483478 27289101 21292972 25244985 18464734 18563158
21719571 20798317 18362917 19820115 26227816 24508170 28004739 30249741 21282665 28648823 24845553 20075255 23537049 17994087 15632085 25830018 20132446 21364917 10731132 12537572 26108605 25315136 26999592 17510324 20966253 12364791 26483478 27289101 21292972 25244985 18464734 18563158
EMBL
BABH01034221
KZ149945
PZC76813.1
NWSH01002867
PCG67417.1
KQ459604
+ More
KPI92112.1 ODYU01008185 SOQ51572.1 RSAL01000004 RVE54549.1 AGBW02011505 OWR46547.1 KQ460205 KPJ17033.1 GEDC01021951 JAS15347.1 PYGN01001024 PSN38358.1 KQ982845 KYQ49989.1 KQ978317 KYM95249.1 ADTU01017434 KQ980804 KYN12769.1 GL887813 EGI69978.1 KQ976453 KYM85469.1 GBYB01006715 JAG76482.1 GL434548 EFN74783.1 KQ971354 EFA06892.2 GECU01012316 JAS95390.1 GL448708 EFN84014.1 JTDY01009882 KOB62369.1 GECZ01006323 JAS63446.1 KK107746 EZA47943.1 GEZM01017039 JAV90946.1 QOIP01000010 RLU18099.1 GL765502 EFZ16193.1 GEBQ01029457 JAT10520.1 ABLF02029984 NNAY01000228 OXU29872.1 KK852510 KDR22332.1 KQ981920 KYN33034.1 QCYY01000951 ROT81595.1 GECZ01028401 JAS41368.1 KQ435736 KOX77080.1 APGK01025659 APGK01025660 KB740600 ENN80090.1 CH933806 KRG02113.1 LBMM01000467 KMQ98266.1 CH916373 CH964232 GDRN01107375 JAI57421.1 CM000070 CH479186 CH902617 CP012526 ALC46089.1 OUUW01000005 SPP80753.1 CVRI01000066 CRL06003.1 KQ414894 KOC59750.1 GEZM01017040 JAV90944.1 GDHF01028685 JAI23629.1 CH954182 GBXI01012635 GBXI01008381 JAD01657.1 JAD05911.1 AB514521 JF309280 AE014297 JRES01001565 KNC22009.1 DS231816 CH940652 BT133496 AFI71926.1 LNIX01000001 OXA62994.1 GDUN01000985 JAN94934.1 GFDL01010985 JAV24060.1 APCN01002504 AXCM01000141 AAAB01008964 GFWV01003064 MAA27794.1 JXUM01070964 KQ562637 KXJ75439.1 JXUM01071241 KQ562651 KXJ75401.1 GDIQ01220837 JAK30888.1 GDIP01017217 JAM86498.1 GDIQ01018632 JAN76105.1 LJIJ01000486 ODM96936.1 RQTK01000965 RUS73209.1 AXCN02000870 LRGB01003123 KZS04507.1 GL732524 EFX89545.1 KZ288305 PBC28769.1 AAPN01106516 AAPN01106517 AAPN01106518 ACPB03009292 GDHF01026821 JAI25493.1 GDIP01139391 JAL64323.1 KQ434809 KZC06617.1 GG666540 EEN57550.1
KPI92112.1 ODYU01008185 SOQ51572.1 RSAL01000004 RVE54549.1 AGBW02011505 OWR46547.1 KQ460205 KPJ17033.1 GEDC01021951 JAS15347.1 PYGN01001024 PSN38358.1 KQ982845 KYQ49989.1 KQ978317 KYM95249.1 ADTU01017434 KQ980804 KYN12769.1 GL887813 EGI69978.1 KQ976453 KYM85469.1 GBYB01006715 JAG76482.1 GL434548 EFN74783.1 KQ971354 EFA06892.2 GECU01012316 JAS95390.1 GL448708 EFN84014.1 JTDY01009882 KOB62369.1 GECZ01006323 JAS63446.1 KK107746 EZA47943.1 GEZM01017039 JAV90946.1 QOIP01000010 RLU18099.1 GL765502 EFZ16193.1 GEBQ01029457 JAT10520.1 ABLF02029984 NNAY01000228 OXU29872.1 KK852510 KDR22332.1 KQ981920 KYN33034.1 QCYY01000951 ROT81595.1 GECZ01028401 JAS41368.1 KQ435736 KOX77080.1 APGK01025659 APGK01025660 KB740600 ENN80090.1 CH933806 KRG02113.1 LBMM01000467 KMQ98266.1 CH916373 CH964232 GDRN01107375 JAI57421.1 CM000070 CH479186 CH902617 CP012526 ALC46089.1 OUUW01000005 SPP80753.1 CVRI01000066 CRL06003.1 KQ414894 KOC59750.1 GEZM01017040 JAV90944.1 GDHF01028685 JAI23629.1 CH954182 GBXI01012635 GBXI01008381 JAD01657.1 JAD05911.1 AB514521 JF309280 AE014297 JRES01001565 KNC22009.1 DS231816 CH940652 BT133496 AFI71926.1 LNIX01000001 OXA62994.1 GDUN01000985 JAN94934.1 GFDL01010985 JAV24060.1 APCN01002504 AXCM01000141 AAAB01008964 GFWV01003064 MAA27794.1 JXUM01070964 KQ562637 KXJ75439.1 JXUM01071241 KQ562651 KXJ75401.1 GDIQ01220837 JAK30888.1 GDIP01017217 JAM86498.1 GDIQ01018632 JAN76105.1 LJIJ01000486 ODM96936.1 RQTK01000965 RUS73209.1 AXCN02000870 LRGB01003123 KZS04507.1 GL732524 EFX89545.1 KZ288305 PBC28769.1 AAPN01106516 AAPN01106517 AAPN01106518 ACPB03009292 GDHF01026821 JAI25493.1 GDIP01139391 JAL64323.1 KQ434809 KZC06617.1 GG666540 EEN57550.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000283053
UP000007151
UP000053240
+ More
UP000245037 UP000075809 UP000078542 UP000005205 UP000078492 UP000007755 UP000078540 UP000000311 UP000007266 UP000008237 UP000037510 UP000053097 UP000279307 UP000007819 UP000215335 UP000005203 UP000027135 UP000078541 UP000002358 UP000283509 UP000053105 UP000019118 UP000009192 UP000036403 UP000001070 UP000007798 UP000001819 UP000008744 UP000007801 UP000092553 UP000078200 UP000268350 UP000092445 UP000092443 UP000183832 UP000053825 UP000192221 UP000091820 UP000008711 UP000000803 UP000037069 UP000095301 UP000002320 UP000008792 UP000198287 UP000095300 UP000075902 UP000076407 UP000075840 UP000075883 UP000075882 UP000075903 UP000069940 UP000249989 UP000075920 UP000075901 UP000075884 UP000094527 UP000271974 UP000075886 UP000076858 UP000075900 UP000000305 UP000076408 UP000242457 UP000002279 UP000015103 UP000076502 UP000001554
UP000245037 UP000075809 UP000078542 UP000005205 UP000078492 UP000007755 UP000078540 UP000000311 UP000007266 UP000008237 UP000037510 UP000053097 UP000279307 UP000007819 UP000215335 UP000005203 UP000027135 UP000078541 UP000002358 UP000283509 UP000053105 UP000019118 UP000009192 UP000036403 UP000001070 UP000007798 UP000001819 UP000008744 UP000007801 UP000092553 UP000078200 UP000268350 UP000092445 UP000092443 UP000183832 UP000053825 UP000192221 UP000091820 UP000008711 UP000000803 UP000037069 UP000095301 UP000002320 UP000008792 UP000198287 UP000095300 UP000075902 UP000076407 UP000075840 UP000075883 UP000075882 UP000075903 UP000069940 UP000249989 UP000075920 UP000075901 UP000075884 UP000094527 UP000271974 UP000075886 UP000076858 UP000075900 UP000000305 UP000076408 UP000242457 UP000002279 UP000015103 UP000076502 UP000001554
Pfam
Interpro
IPR041095
EFG_II
+ More
IPR009000 Transl_B-barrel_sf
IPR035649 EFG_V
IPR027417 P-loop_NTPase
IPR035647 EFG_III/V
IPR020568 Ribosomal_S5_D2-typ_fold
IPR031157 G_TR_CS
IPR000795 TF_GTP-bd_dom
IPR009022 EFG_III
IPR005225 Small_GTP-bd_dom
IPR000640 EFG_V-like
IPR014721 Ribosomal_S5_D2-typ_fold_subgr
IPR004161 EFTu-like_2
IPR005517 Transl_elong_EFG/EF2_IV
IPR001648 Ribosomal_S18
IPR036870 Ribosomal_S18_sf
IPR030851 EFG2
IPR038495 ATPase_E_C
IPR002842 ATPase_V1_Esu
IPR009000 Transl_B-barrel_sf
IPR035649 EFG_V
IPR027417 P-loop_NTPase
IPR035647 EFG_III/V
IPR020568 Ribosomal_S5_D2-typ_fold
IPR031157 G_TR_CS
IPR000795 TF_GTP-bd_dom
IPR009022 EFG_III
IPR005225 Small_GTP-bd_dom
IPR000640 EFG_V-like
IPR014721 Ribosomal_S5_D2-typ_fold_subgr
IPR004161 EFTu-like_2
IPR005517 Transl_elong_EFG/EF2_IV
IPR001648 Ribosomal_S18
IPR036870 Ribosomal_S18_sf
IPR030851 EFG2
IPR038495 ATPase_E_C
IPR002842 ATPase_V1_Esu
SUPFAM
Gene 3D
ProteinModelPortal
H9IU35
A0A2W1BYG6
A0A2A4J5K6
A0A194PFP7
A0A2H1WFC6
A0A3S2M9K1
+ More
A0A212EYL2 A0A194RGT7 A0A1B6CPE1 A0A2P8Y271 A0A151WQ12 A0A195C310 A0A158NJ41 A0A195DJU9 F4W794 A0A195BL22 A0A0C9REE6 E1ZVL4 D6WPX8 A0A1B6J8A0 E2BK05 A0A0L7KHK7 A0A1B6GLX8 A0A026VVW9 A0A1Y1N3N6 A0A3L8DDX8 E9IT92 A0A1B6KGD5 J9JKG8 A0A232FGQ4 A0A087ZT96 A0A067REY6 A0A195EYS5 K7J309 A0A3R7QK99 A0A1B6EU19 A0A0M9A4I1 N6UEF1 A0A0Q9XCL1 A0A0J7L6I1 B4JSI3 B4NAU8 A0A0P4W9E1 Q29BD5 B4GNT0 B3M011 A0A0M3QXL7 A0A1A9VVW3 A0A3B0JJ22 A0A1A9Z3R1 A0A1A9XFK6 A0A1J1J0F0 A0A0L7QMD6 A0A1Y1MZ82 A0A1W4W1J0 A0A1A9WRW7 A0A0K8UAG6 B3P8M3 A0A0A1X4W8 Q9VCX4 A0A0L0BS17 A0A1I8MHD4 B0W010 B4M416 I1WYH6 A0A226F0P9 A0A0P6IYK0 A0A1S4FZT8 A0A1Q3F921 A0A1I8PK89 A0A182TL00 A0A182WTC1 A0A182HVB4 A0A182M335 A0A182L264 A0A182VKM0 A0A1S4GWL9 A0A293LH03 A0A182GKZ0 A0A182GL48 A0A0P5HXK6 A0A0P6BC28 A0A182W9S8 A0A0P6IQC6 A0A182ST14 A0A182NE67 A0A1D2MVA9 A0A3S1BRK1 A0A182QEH8 A0A164LWQ0 A0A182RBQ2 E9FU13 A0A182YGX6 A0A2A3EC44 F7G7T5 T1HV44 A0A0K8UG02 A0A0N8CEK7 A0A154P443 C3YQ46
A0A212EYL2 A0A194RGT7 A0A1B6CPE1 A0A2P8Y271 A0A151WQ12 A0A195C310 A0A158NJ41 A0A195DJU9 F4W794 A0A195BL22 A0A0C9REE6 E1ZVL4 D6WPX8 A0A1B6J8A0 E2BK05 A0A0L7KHK7 A0A1B6GLX8 A0A026VVW9 A0A1Y1N3N6 A0A3L8DDX8 E9IT92 A0A1B6KGD5 J9JKG8 A0A232FGQ4 A0A087ZT96 A0A067REY6 A0A195EYS5 K7J309 A0A3R7QK99 A0A1B6EU19 A0A0M9A4I1 N6UEF1 A0A0Q9XCL1 A0A0J7L6I1 B4JSI3 B4NAU8 A0A0P4W9E1 Q29BD5 B4GNT0 B3M011 A0A0M3QXL7 A0A1A9VVW3 A0A3B0JJ22 A0A1A9Z3R1 A0A1A9XFK6 A0A1J1J0F0 A0A0L7QMD6 A0A1Y1MZ82 A0A1W4W1J0 A0A1A9WRW7 A0A0K8UAG6 B3P8M3 A0A0A1X4W8 Q9VCX4 A0A0L0BS17 A0A1I8MHD4 B0W010 B4M416 I1WYH6 A0A226F0P9 A0A0P6IYK0 A0A1S4FZT8 A0A1Q3F921 A0A1I8PK89 A0A182TL00 A0A182WTC1 A0A182HVB4 A0A182M335 A0A182L264 A0A182VKM0 A0A1S4GWL9 A0A293LH03 A0A182GKZ0 A0A182GL48 A0A0P5HXK6 A0A0P6BC28 A0A182W9S8 A0A0P6IQC6 A0A182ST14 A0A182NE67 A0A1D2MVA9 A0A3S1BRK1 A0A182QEH8 A0A164LWQ0 A0A182RBQ2 E9FU13 A0A182YGX6 A0A2A3EC44 F7G7T5 T1HV44 A0A0K8UG02 A0A0N8CEK7 A0A154P443 C3YQ46
PDB
6NOT
E-value=1.58234e-115,
Score=1067
Ontologies
GO
Topology
Subcellular location
Mitochondrion
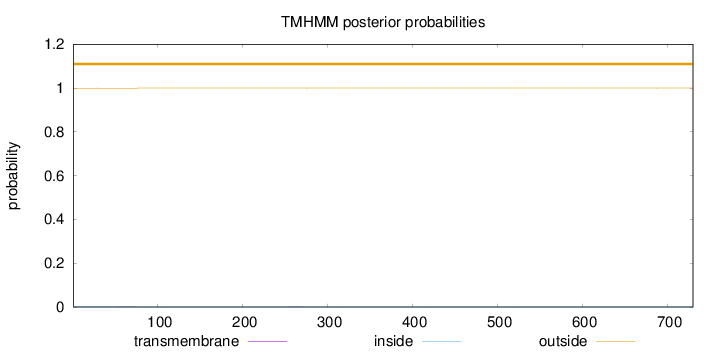
Length:
730
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01229
Exp number, first 60 AAs:
0.00282
Total prob of N-in:
0.00066
outside
1 - 730
Population Genetic Test Statistics
Pi
267.401805
Theta
178.897439
Tajima's D
1.684151
CLR
0.324704
CSRT
0.822558872056397
Interpretation
Uncertain
