Gene
KWMTBOMO14940
Pre Gene Modal
BGIBMGA006890
Annotation
PREDICTED:_uncharacterized_protein_K02A2.6-like_isoform_X1_[Amyelois_transitella]
Location in the cell
Nuclear Reliability : 3.765
Sequence
CDS
ATGGCAACAAATAACAACGAAGCAGTCTCCGGTATTAAACCACCCGGATACTTGCACATCGAGTCGGACAACAAGTCGGCAAATTGGAAAAAATGGCGACAACAGTTCGAATGGTATGCAACAGCAATTCAGCTAGGAAAGAAACCTGCCGATATTCAAGCTGCAACATTCATGTCAATCATAGGACCAGATGCAATTGACATCTACAACAGTTTTAATTTGAATACAATAGACGAACAAAAATTAAGCATCATTATCGGAAAGTTCGAAGAATATTTCGCGCCGAAAAACAACATATCGTTCGAAAGGTATGTTTTCTTCAAAATTGAACAACATGAAGACGAATCGTTTAACGAATTCATCACTAGAATTAAAACACAAGCAAACAAATGCGAGTTTGGCACTCTACTTGAAGAAATGCTAAAAGACAAGATTGTATTCGGAATAAAATCAACCCAAATCAGAGAAAAATTACTCACCGAAGAAAAACTTGATCTCACAAAAGCCACAGCCATCTGTAAAAGCAGTGAACAAGCTTCCAAACAACTTAACGAGTTTGAGAATAACAACAAGAGTGAAAAAATACTGACAATCAAAAGTAAGAACTTCAGAAATGAAAAATTTGACTGTAAAAGATGTGGTTCAAGCCACAAGGCTAGAGAATGCCAAGCCTACAACAAATTATGCACAAAGTGTAACAAGGCCGGTCATTTTGCAAAAATGTGTCGTTCCCAAATCCAATTGAAACAAAAAAATAAAATAAATACTCTAGAAGAAAATTCTACTTCCGAAAATTCAGATGAATCATTTATAGGACAATTAAGTGTCGGCAACTCCAAAGATTGGACAGAAAGTGTCGAAACTGCCAACACCAAATTCACAGCAAAGCTTGATACTGGTGCAGAGTGCAACGTCCTTCCAAATTTCATAGTACAGAAAACTCATGCTTGTATACAACCAAGTCGCACTAAAAATTTAATAAGTTATTCAAATAACAGAATTTCAGTACTTGGAGAAGTAAAGTTACAATGCAAAATAAAAAAGAAAACTGCTAAAATACTATTTAAAGTTGTAGCTGAAAGAGTCACACCAGTCTTAGGACTAAACACTTGTGAGAAGCTGGGACTAATTGCACGAGTTGAAACATTGAAAGAAAGCTCATCCAATGATGACATATTTAAAGGGTTAGGTTGCTACAAAAATTTTGAGTATGATATTGACCTCATAGAAAACCCCAAATTTGAAATAAAACCAACAAGAAAAGTTCCACATGCCATAAGAAATGAAGTAAAACAAGAATTAGATAGAATGGTAAAATTGGATGTCATCACACCTGAAACAGAACCGACACCAGCAGTAAGTCCGATGGTCGTAGTCAGACAAAAAGGAAAATTGAGAATATGCATCGACCCATCGGATGTGAACAAAAATATTTTAAGAAGACATTTCCCACTTGCAACAATAGAAGAGATAGCAGCAGACATCAAAGGATCAGAATACTTTTCCCTACTAGACTGCACCAAAGGATTTTGGCAAATTAAATTATCAAAAAGAACACAAAAGATACTGACATTTTCTACACCATGGGGAAGATATTCATTCAAAAGACTTCCATTCGGACTATCGTCAGCCCCTGAAATTTTTCAAGAAATCTTGACTAATCTTCTCAGCAAATTCAAAAATGTAAAAGTATCAATCGATGATATCTTTATTCATGCCAAAAAGAAGGAAGAGCTACAGAAAACGGTCAAAGAAGTAATAGAGACATTGAAAATTTCTGGCTTCAAACTCAATCAAAGCAAATGCATCTTTGAAGCCAAAAGAATAAAATTTTTGGGTCATATAGTCTCAGCCAAAGGTTTAGAAGCAGATCCTGAAAAAGTCAAAGCCATAGAATTTATGAAAACACCACAGAACAAAAAAGAACTGCAAAGGATACTAGGAATGATAACATATTTAAATAAATACATACCCAACATGTCAGAATTAACAAACCCTTTGAGAGACCTACTTCACAAGGACACAAGTTTCAACTGGGAATTCTATCATGATGAAGCACTCAACAAAATTAAAAAAGTTTTACAGAATCCGCCAGTGCTAAAACTATATGATGTAAACAAACCAGTCACACTAAGCGTGGATGCAAGCTCAAAGAATCTCGGTGCAGCACTTCTTCAGGAAGGCCAACCAGTAGCTTATGGAGCCAGAACGTTGACTAAATCCGAACAAAATTATCCTCAAATTGAAAAGGAAGCACTTGCTATACTATTTGGGTGCAAAAAGTTCCATGAGTATGTCTATGGCAAAGAGTTATTAGTCGAATCAGATCACAAGCCGCTCGAGACAATATTCAGAAAAAACATTCAGTCAGCCCCAGCTAGATTTCAACGTATAATGCTCAACCTAACACCGTATTCACCAAAAATTACATTTAAAAAAGGCACAGAAATACCACTAGCAGACTTTTTAAGTCGTGATTGTGACACTTCAAAACTGAATGACTATCAGGAAGAAGACCTGAAGATCTCAATAATTTTACCAACAACATTTGACTACACAGAAGAGTTGATCAAAGCAACAAAAGAAGACCCCCACCTGCAACTCTTGTTAAAAACAATAATGAAAGGGTTCCCAGAAAAGGCTGATCAACTCCCTACAGAACTGCATCACTACTTCAATTTCAAAGAAGAACTAACATACTTCAAAGGTCTAATTTTCAAAGGTCACAAAATTGTAGTGCCCAAAACACAAATACCTCAGATGCTAAAATACATACACCAAGGTCACCTTGGAATTCAGAGTTGCTTGAAAAGAGCACACCAATTACTCTATTGGAGAGGACAATATGAAGACATTGTGAAACTGGTTAAAAGCTGCTCAGCATGTGAAAAAACACAAAAGGACAATACCAATTATACCGTAGCAGTCAAAAGAATCCCATCTCTCCCATGGCAGATTGTGGCATCTGATTTATTTGAACTAAAAGGAAAACAGTATATTAAAGAATGGCGTTTCAAGCACTCAACATCCAGTCCTCACCACCCCCAAGGCAATGGCTTAGCCGAAAGAGCTGTCCAAACTGCAAAAAACCTACTAAGGAAATGCAGTATTGATAAATCGGATATCCAACTTGCTCTATTAAATTGGAGAAATACACCAAGAACAAACAATCTAGGGTCTCCTAATCAACGCCTCTACAGTAGAATAACACGATCATTAATACCTACAAGTGAAAATAATCTTAAACCAAAGATACTTCAAGGGGTCACATCCGAACTAAAGTACCTACGAAACAAACAAGCCAACCATAGCAACAAGCAACGTAAGGAGCCAACCAATTATGAACTCGACGACAGAATCAGACACAAAGTCGGTCATCGTCAATGGGAAGGAGCGAGAGTGATAGAAAAACCTAATAATCACCCGAGATCCATTATCATTAAAACTGACAAAGGACAAATATTCAGAAGAAACATTGGACACATACACAATACACTATCAGACCTATCAATTGGCAGCAAAGAAGCGGTTCCGGAAACAGTATATCCTGACGACTATCCACATGCCCAATCGGACAAACAACCGGAGCTACCTGCAGCAGCTACTACATCTCAAGAACAAACAACTATATCACCAAGGATTAATTGCAAAAGCAGCGCTTCAGAAGCACCCGTCATAACTAGCAGTGATCAACCGACTACAAGATGTTCAAGATTCGGAAGAACCATCAAACCAGTCGACAGACTAGATCTATAG
Protein
MATNNNEAVSGIKPPGYLHIESDNKSANWKKWRQQFEWYATAIQLGKKPADIQAATFMSIIGPDAIDIYNSFNLNTIDEQKLSIIIGKFEEYFAPKNNISFERYVFFKIEQHEDESFNEFITRIKTQANKCEFGTLLEEMLKDKIVFGIKSTQIREKLLTEEKLDLTKATAICKSSEQASKQLNEFENNNKSEKILTIKSKNFRNEKFDCKRCGSSHKARECQAYNKLCTKCNKAGHFAKMCRSQIQLKQKNKINTLEENSTSENSDESFIGQLSVGNSKDWTESVETANTKFTAKLDTGAECNVLPNFIVQKTHACIQPSRTKNLISYSNNRISVLGEVKLQCKIKKKTAKILFKVVAERVTPVLGLNTCEKLGLIARVETLKESSSNDDIFKGLGCYKNFEYDIDLIENPKFEIKPTRKVPHAIRNEVKQELDRMVKLDVITPETEPTPAVSPMVVVRQKGKLRICIDPSDVNKNILRRHFPLATIEEIAADIKGSEYFSLLDCTKGFWQIKLSKRTQKILTFSTPWGRYSFKRLPFGLSSAPEIFQEILTNLLSKFKNVKVSIDDIFIHAKKKEELQKTVKEVIETLKISGFKLNQSKCIFEAKRIKFLGHIVSAKGLEADPEKVKAIEFMKTPQNKKELQRILGMITYLNKYIPNMSELTNPLRDLLHKDTSFNWEFYHDEALNKIKKVLQNPPVLKLYDVNKPVTLSVDASSKNLGAALLQEGQPVAYGARTLTKSEQNYPQIEKEALAILFGCKKFHEYVYGKELLVESDHKPLETIFRKNIQSAPARFQRIMLNLTPYSPKITFKKGTEIPLADFLSRDCDTSKLNDYQEEDLKISIILPTTFDYTEELIKATKEDPHLQLLLKTIMKGFPEKADQLPTELHHYFNFKEELTYFKGLIFKGHKIVVPKTQIPQMLKYIHQGHLGIQSCLKRAHQLLYWRGQYEDIVKLVKSCSACEKTQKDNTNYTVAVKRIPSLPWQIVASDLFELKGKQYIKEWRFKHSTSSPHHPQGNGLAERAVQTAKNLLRKCSIDKSDIQLALLNWRNTPRTNNLGSPNQRLYSRITRSLIPTSENNLKPKILQGVTSELKYLRNKQANHSNKQRKEPTNYELDDRIRHKVGHRQWEGARVIEKPNNHPRSIIIKTDKGQIFRRNIGHIHNTLSDLSIGSKEAVPETVYPDDYPHAQSDKQPELPAAATTSQEQTTISPRINCKSSASEAPVITSSDQPTTRCSRFGRTIKPVDRLDL
Summary
Uniprot
A0A3P8Q6C6
A0A3B3HLX7
A0A2G8KFU3
A0A2G8KHQ8
A0A3B1JKR1
A0A2G8LMB1
+ More
A0A3P9HT22 A0A3B3C9X5 A0A3P9HXP1 A0A1A8UGA0 A0A1A8DQM9 A0A147BKH7 A0A147BJX4 A0A0A9Z4Y4 A0A1S3JEC7 A0A3P8P6K4 A0A146LC45 A0A1A8S1T9 A0A069DZS8 A0A069DY08 A0A146TAZ6 J9L288 A0A3P9K9H2 A0A3P8RKT7 A0A3P9LZF9 A0A3P9AYU2 X1WTY2 W4YGV7 A0A3B3H7E4 W4Y1E4 A0A1Y1M671 A0A1S3HHB2 A0A3P8QAD1 A0A0P4XRP4 A0A0P5V4F7 A0A0P5A948 A0A0P6BAV5 A0A0P5VCW1 A0A146L0H7 A0A1S3DNR0 A0A1S3DH71 A0A1Y1NDC5 A0A1S3DM37 A0A2S2NI46 A0A0P5DHJ0 A0A3P9L920 W4XLW6 A0A3P8Q2Q2 A0A075EIL5 W4YP04 A0A0P5VRF2 A0A3B3HBT8 W4ZHL7 A0A0P5XDK7 A0A3P8RHF8 A0A087SXG5 A0A0P5UUP4 A0A3P8QB87 A0A0P6BCI0 A0A3Q0JFZ1 A0A3Q0JJE3 A0A147BN28 A0A1Y1L7V1 A0A3Q2XZ76 A0A1Y1MCJ7 A0A1S3DJN8 A0A3P8S414 A0A023EY30 A0A3B1ITQ0 A0A2G8JDW4 A0A1Y1KVT8 A0A1A8L2U7 A0A1Y1NHV7 A0A3B3I1S4 A0A1A8QJ23 A0A147BIU1 A0A0P4VZQ4 A0A0P5UT77 A0A0A9XW98 A0A3B3IIM2 A0A0P5TYX3 A0A1A8AMK6 A0A3Q0JK14 A0A0P5C1M8 A0A3Q2DE46 A0A023EXW7 A0A3P8QUW8 A0A087UXZ8 W4ZAI4 A0A3B1K366 A0A147BIE4 A0A023EWW4 A0A0A9WHB1 A0A1S3DLU0 A0A1S3DMQ1 A0A0V0G5U8 A0A0P4ZSZ7
A0A3P9HT22 A0A3B3C9X5 A0A3P9HXP1 A0A1A8UGA0 A0A1A8DQM9 A0A147BKH7 A0A147BJX4 A0A0A9Z4Y4 A0A1S3JEC7 A0A3P8P6K4 A0A146LC45 A0A1A8S1T9 A0A069DZS8 A0A069DY08 A0A146TAZ6 J9L288 A0A3P9K9H2 A0A3P8RKT7 A0A3P9LZF9 A0A3P9AYU2 X1WTY2 W4YGV7 A0A3B3H7E4 W4Y1E4 A0A1Y1M671 A0A1S3HHB2 A0A3P8QAD1 A0A0P4XRP4 A0A0P5V4F7 A0A0P5A948 A0A0P6BAV5 A0A0P5VCW1 A0A146L0H7 A0A1S3DNR0 A0A1S3DH71 A0A1Y1NDC5 A0A1S3DM37 A0A2S2NI46 A0A0P5DHJ0 A0A3P9L920 W4XLW6 A0A3P8Q2Q2 A0A075EIL5 W4YP04 A0A0P5VRF2 A0A3B3HBT8 W4ZHL7 A0A0P5XDK7 A0A3P8RHF8 A0A087SXG5 A0A0P5UUP4 A0A3P8QB87 A0A0P6BCI0 A0A3Q0JFZ1 A0A3Q0JJE3 A0A147BN28 A0A1Y1L7V1 A0A3Q2XZ76 A0A1Y1MCJ7 A0A1S3DJN8 A0A3P8S414 A0A023EY30 A0A3B1ITQ0 A0A2G8JDW4 A0A1Y1KVT8 A0A1A8L2U7 A0A1Y1NHV7 A0A3B3I1S4 A0A1A8QJ23 A0A147BIU1 A0A0P4VZQ4 A0A0P5UT77 A0A0A9XW98 A0A3B3IIM2 A0A0P5TYX3 A0A1A8AMK6 A0A3Q0JK14 A0A0P5C1M8 A0A3Q2DE46 A0A023EXW7 A0A3P8QUW8 A0A087UXZ8 W4ZAI4 A0A3B1K366 A0A147BIE4 A0A023EWW4 A0A0A9WHB1 A0A1S3DLU0 A0A1S3DMQ1 A0A0V0G5U8 A0A0P4ZSZ7
Pubmed
EMBL
MRZV01000618
PIK46835.1
MRZV01000576
PIK47528.1
MRZV01000033
PIK61397.1
+ More
HADY01023773 HAEJ01006887 SBS47344.1 HADZ01002490 HAEA01007842 SBQ36322.1 GEGO01004116 JAR91288.1 GEGO01004317 JAR91087.1 GBHO01004090 JAG39514.1 GDHC01013957 JAQ04672.1 HAEH01020882 SBS11479.1 GBGD01000115 JAC88774.1 GBGD01000119 JAC88770.1 GCES01096219 JAQ90103.1 ABLF02022355 ABLF02022359 ABLF02022361 ABLF02022365 ABLF02041467 ABLF02013930 ABLF02013934 AAGJ04164197 AAGJ04066141 GEZM01041944 JAV80090.1 GDIP01239061 JAI84340.1 GDIP01199810 GDIP01181260 GDIP01124130 GDIP01104931 GDIP01053137 GDIP01051184 JAL98783.1 GDIP01203362 JAJ20040.1 GDIP01019161 JAM84554.1 GDIP01102077 JAM01638.1 GDHC01017753 JAQ00876.1 GEZM01011167 JAV93617.1 GGMR01004179 MBY16798.1 GDIP01156216 JAJ67186.1 AAGJ04172695 KF319019 AIE48224.1 AAGJ04126635 GDIP01096578 JAM07137.1 AAGJ04068764 GDIP01073678 JAM30037.1 KK112400 KFM57554.1 GDIP01175260 GDIP01109876 JAL93838.1 GDIP01017210 JAM86505.1 GEGO01003204 JAR92200.1 GEZM01066276 JAV68015.1 GEZM01035137 JAV83383.1 GBBI01004745 JAC13967.1 MRZV01002352 PIK33923.1 GEZM01074317 JAV64601.1 HAEF01001580 SBR38962.1 GEZM01001973 JAV97431.1 HAEH01011921 SBR93481.1 GEGO01004733 JAR90671.1 GDKW01000526 JAI56069.1 GDIP01175266 GDIP01109877 JAL93837.1 GBHO01022049 GBHO01022048 JAG21555.1 JAG21556.1 GDIP01122420 JAL81294.1 HADY01017769 SBP56254.1 GDIP01192261 JAJ31141.1 GBBI01004851 JAC13861.1 KK122208 KFM82237.1 AAGJ04077500 GEGO01004841 JAR90563.1 GBBI01004852 JAC13860.1 GBHO01036465 JAG07139.1 GECL01002751 JAP03373.1 GDIP01210008 JAJ13394.1
HADY01023773 HAEJ01006887 SBS47344.1 HADZ01002490 HAEA01007842 SBQ36322.1 GEGO01004116 JAR91288.1 GEGO01004317 JAR91087.1 GBHO01004090 JAG39514.1 GDHC01013957 JAQ04672.1 HAEH01020882 SBS11479.1 GBGD01000115 JAC88774.1 GBGD01000119 JAC88770.1 GCES01096219 JAQ90103.1 ABLF02022355 ABLF02022359 ABLF02022361 ABLF02022365 ABLF02041467 ABLF02013930 ABLF02013934 AAGJ04164197 AAGJ04066141 GEZM01041944 JAV80090.1 GDIP01239061 JAI84340.1 GDIP01199810 GDIP01181260 GDIP01124130 GDIP01104931 GDIP01053137 GDIP01051184 JAL98783.1 GDIP01203362 JAJ20040.1 GDIP01019161 JAM84554.1 GDIP01102077 JAM01638.1 GDHC01017753 JAQ00876.1 GEZM01011167 JAV93617.1 GGMR01004179 MBY16798.1 GDIP01156216 JAJ67186.1 AAGJ04172695 KF319019 AIE48224.1 AAGJ04126635 GDIP01096578 JAM07137.1 AAGJ04068764 GDIP01073678 JAM30037.1 KK112400 KFM57554.1 GDIP01175260 GDIP01109876 JAL93838.1 GDIP01017210 JAM86505.1 GEGO01003204 JAR92200.1 GEZM01066276 JAV68015.1 GEZM01035137 JAV83383.1 GBBI01004745 JAC13967.1 MRZV01002352 PIK33923.1 GEZM01074317 JAV64601.1 HAEF01001580 SBR38962.1 GEZM01001973 JAV97431.1 HAEH01011921 SBR93481.1 GEGO01004733 JAR90671.1 GDKW01000526 JAI56069.1 GDIP01175266 GDIP01109877 JAL93837.1 GBHO01022049 GBHO01022048 JAG21555.1 JAG21556.1 GDIP01122420 JAL81294.1 HADY01017769 SBP56254.1 GDIP01192261 JAJ31141.1 GBBI01004851 JAC13861.1 KK122208 KFM82237.1 AAGJ04077500 GEGO01004841 JAR90563.1 GBBI01004852 JAC13860.1 GBHO01036465 JAG07139.1 GECL01002751 JAP03373.1 GDIP01210008 JAJ13394.1
Proteomes
Pfam
Interpro
IPR001584
Integrase_cat-core
+ More
IPR041577 RT_RNaseH_2
IPR001878 Znf_CCHC
IPR012337 RNaseH-like_sf
IPR021109 Peptidase_aspartic_dom_sf
IPR000477 RT_dom
IPR041588 Integrase_H2C2
IPR036397 RNaseH_sf
IPR001995 Peptidase_A2_cat
IPR036875 Znf_CCHC_sf
IPR005162 Retrotrans_gag_dom
IPR001969 Aspartic_peptidase_AS
IPR041577 RT_RNaseH_2
IPR001878 Znf_CCHC
IPR012337 RNaseH-like_sf
IPR021109 Peptidase_aspartic_dom_sf
IPR000477 RT_dom
IPR041588 Integrase_H2C2
IPR036397 RNaseH_sf
IPR001995 Peptidase_A2_cat
IPR036875 Znf_CCHC_sf
IPR005162 Retrotrans_gag_dom
IPR001969 Aspartic_peptidase_AS
Gene 3D
ProteinModelPortal
A0A3P8Q6C6
A0A3B3HLX7
A0A2G8KFU3
A0A2G8KHQ8
A0A3B1JKR1
A0A2G8LMB1
+ More
A0A3P9HT22 A0A3B3C9X5 A0A3P9HXP1 A0A1A8UGA0 A0A1A8DQM9 A0A147BKH7 A0A147BJX4 A0A0A9Z4Y4 A0A1S3JEC7 A0A3P8P6K4 A0A146LC45 A0A1A8S1T9 A0A069DZS8 A0A069DY08 A0A146TAZ6 J9L288 A0A3P9K9H2 A0A3P8RKT7 A0A3P9LZF9 A0A3P9AYU2 X1WTY2 W4YGV7 A0A3B3H7E4 W4Y1E4 A0A1Y1M671 A0A1S3HHB2 A0A3P8QAD1 A0A0P4XRP4 A0A0P5V4F7 A0A0P5A948 A0A0P6BAV5 A0A0P5VCW1 A0A146L0H7 A0A1S3DNR0 A0A1S3DH71 A0A1Y1NDC5 A0A1S3DM37 A0A2S2NI46 A0A0P5DHJ0 A0A3P9L920 W4XLW6 A0A3P8Q2Q2 A0A075EIL5 W4YP04 A0A0P5VRF2 A0A3B3HBT8 W4ZHL7 A0A0P5XDK7 A0A3P8RHF8 A0A087SXG5 A0A0P5UUP4 A0A3P8QB87 A0A0P6BCI0 A0A3Q0JFZ1 A0A3Q0JJE3 A0A147BN28 A0A1Y1L7V1 A0A3Q2XZ76 A0A1Y1MCJ7 A0A1S3DJN8 A0A3P8S414 A0A023EY30 A0A3B1ITQ0 A0A2G8JDW4 A0A1Y1KVT8 A0A1A8L2U7 A0A1Y1NHV7 A0A3B3I1S4 A0A1A8QJ23 A0A147BIU1 A0A0P4VZQ4 A0A0P5UT77 A0A0A9XW98 A0A3B3IIM2 A0A0P5TYX3 A0A1A8AMK6 A0A3Q0JK14 A0A0P5C1M8 A0A3Q2DE46 A0A023EXW7 A0A3P8QUW8 A0A087UXZ8 W4ZAI4 A0A3B1K366 A0A147BIE4 A0A023EWW4 A0A0A9WHB1 A0A1S3DLU0 A0A1S3DMQ1 A0A0V0G5U8 A0A0P4ZSZ7
A0A3P9HT22 A0A3B3C9X5 A0A3P9HXP1 A0A1A8UGA0 A0A1A8DQM9 A0A147BKH7 A0A147BJX4 A0A0A9Z4Y4 A0A1S3JEC7 A0A3P8P6K4 A0A146LC45 A0A1A8S1T9 A0A069DZS8 A0A069DY08 A0A146TAZ6 J9L288 A0A3P9K9H2 A0A3P8RKT7 A0A3P9LZF9 A0A3P9AYU2 X1WTY2 W4YGV7 A0A3B3H7E4 W4Y1E4 A0A1Y1M671 A0A1S3HHB2 A0A3P8QAD1 A0A0P4XRP4 A0A0P5V4F7 A0A0P5A948 A0A0P6BAV5 A0A0P5VCW1 A0A146L0H7 A0A1S3DNR0 A0A1S3DH71 A0A1Y1NDC5 A0A1S3DM37 A0A2S2NI46 A0A0P5DHJ0 A0A3P9L920 W4XLW6 A0A3P8Q2Q2 A0A075EIL5 W4YP04 A0A0P5VRF2 A0A3B3HBT8 W4ZHL7 A0A0P5XDK7 A0A3P8RHF8 A0A087SXG5 A0A0P5UUP4 A0A3P8QB87 A0A0P6BCI0 A0A3Q0JFZ1 A0A3Q0JJE3 A0A147BN28 A0A1Y1L7V1 A0A3Q2XZ76 A0A1Y1MCJ7 A0A1S3DJN8 A0A3P8S414 A0A023EY30 A0A3B1ITQ0 A0A2G8JDW4 A0A1Y1KVT8 A0A1A8L2U7 A0A1Y1NHV7 A0A3B3I1S4 A0A1A8QJ23 A0A147BIU1 A0A0P4VZQ4 A0A0P5UT77 A0A0A9XW98 A0A3B3IIM2 A0A0P5TYX3 A0A1A8AMK6 A0A3Q0JK14 A0A0P5C1M8 A0A3Q2DE46 A0A023EXW7 A0A3P8QUW8 A0A087UXZ8 W4ZAI4 A0A3B1K366 A0A147BIE4 A0A023EWW4 A0A0A9WHB1 A0A1S3DLU0 A0A1S3DMQ1 A0A0V0G5U8 A0A0P4ZSZ7
PDB
4OL8
E-value=7.34022e-49,
Score=494
Ontologies
GO
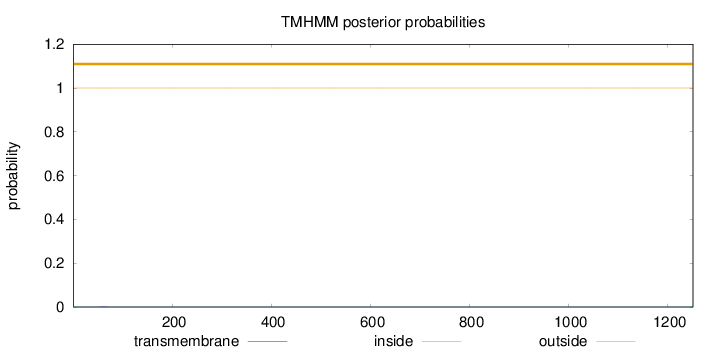
Topology
Length:
1251
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00374
Exp number, first 60 AAs:
0.00173
Total prob of N-in:
0.00018
outside
1 - 1251
Population Genetic Test Statistics
Pi
395.565447
Theta
205.630573
Tajima's D
3.235804
CLR
0.355135
CSRT
0.987200639968002
Interpretation
Uncertain
