Gene
KWMTBOMO14939 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA000804
Annotation
mitochondrial_ATP_synthase_coupling_factor_6_[Bombyx_mori]
Full name
ATP synthase-coupling factor 6, mitochondrial
Location in the cell
Mitochondrial Reliability : 1.701
Sequence
CDS
ATGCTCGTATCAAAACTGGTTGGTTTGAGGGCGGCGACCACTTCCATGATGGTCAGTAGAAATTTAGCGGCAGCTCAGAAAGCTACCGATCCCATCCAACAGCTGTTTTTAGATAAAATCAGGGAATACAAACAGAAGAGCGCGGGTGGCAAAGTCCCAGATGCCAGCCCTGCTGTCGAGAAAGAGCTGAAAACTGAATTAGAAAAGCTTGAGAAGCAGTATGGAGGGGGTCCCGGAATCGACATGACAGCATTCCCATCTCTGAAATTTGAAGAACCTAAGCTGGACCCCATTGATGAACAGGCCGCCCCCAAGAAGTGA
Protein
MLVSKLVGLRAATTSMMVSRNLAAAQKATDPIQQLFLDKIREYKQKSAGGKVPDASPAVEKELKTELEKLEKQYGGGPGIDMTAFPSLKFEEPKLDPIDEQAAPKK
Summary
Description
Mitochondrial membrane ATP synthase (F(1)F(0) ATP synthase or Complex V) produces ATP from ADP in the presence of a proton gradient across the membrane which is generated by electron transport complexes of the respiratory chain.
Similarity
Belongs to the eukaryotic ATPase subunit F6 family.
Belongs to the WD repeat SEC13 family.
Belongs to the WD repeat SEC13 family.
Uniprot
Q1HPM6
A0A0L7KNK4
A0A194PGG3
A0A194RI86
A0A0K8SQL0
A0A2W1BVJ1
+ More
A0A3S2LVB0 A0A212EHB4 A0A2A4JHJ2 A0A2H1W3L0 E3UPD0 A0A2W1BGQ5 A0A2A4JAN6 A0A1J1I514 B8XY08 E2AGN2 Q17H51 A0A1S4F2T8 B6DDV7 A0A182PSQ5 U5EUA2 A0A0J7L2W9 A0A151X7I3 T1E3Q7 A0A195CBQ6 T1DEZ9 A0A2M4AA23 A0A224Y2F2 A0A0T6B055 A0A195DLY5 T1GY90 A0A182XDQ5 A0A1B6C1C8 A0A2M3Z030 A0A182FP58 A0A2M4C473 A0A0P4VHT1 R4FLH7 A0A182LNN7 Q7Q7P4 A0A182IDG4 A0A2P8YYB5 Q5MIP9 T1DFP1 E9IXS2 A0A0V0G3G2 F4X1G8 A0A1B6GI34 A0A069DNC6 A0A182TQ12 A0A182V2I8 A0A1W4XIN9 A0A2J7R4R0 A0A182YET2 D1FQ50 A0A182Q6H2 T1E347 A0A182NDY9 A0A182VPR0 A0A1B6K417 V5GW68 D6WSX9 A0A3L8D6R6 A0A067R407 A0A1B6IK82 A0A0A9XJP8 A0A026WRA3 A0A146KW89 A0A182LXT4 A0A182REB0 A0A182JXH1 A0A182J482 T1J6P0 A0A1B0GQC8 A0A0K8TQT7 A0A1B6KKT7 A0A146M9C8 B4JI06 A0A1B6MGM3 B0WNE9 A0A2H1W5U6 A0A232EX76 K7J3X9 A0A195BJ79 A0A158NA73 B4K4I0 N6UHM1 A0A2R7VZE8 A0A195FRC5 A0A0P4WD74 A0A1L8E016 A0A1L8EF44 R4WIQ2 A0A1L8EF88 B4LVY8 A0A1L8E0A0 A0A336L460 W8BWU4
A0A3S2LVB0 A0A212EHB4 A0A2A4JHJ2 A0A2H1W3L0 E3UPD0 A0A2W1BGQ5 A0A2A4JAN6 A0A1J1I514 B8XY08 E2AGN2 Q17H51 A0A1S4F2T8 B6DDV7 A0A182PSQ5 U5EUA2 A0A0J7L2W9 A0A151X7I3 T1E3Q7 A0A195CBQ6 T1DEZ9 A0A2M4AA23 A0A224Y2F2 A0A0T6B055 A0A195DLY5 T1GY90 A0A182XDQ5 A0A1B6C1C8 A0A2M3Z030 A0A182FP58 A0A2M4C473 A0A0P4VHT1 R4FLH7 A0A182LNN7 Q7Q7P4 A0A182IDG4 A0A2P8YYB5 Q5MIP9 T1DFP1 E9IXS2 A0A0V0G3G2 F4X1G8 A0A1B6GI34 A0A069DNC6 A0A182TQ12 A0A182V2I8 A0A1W4XIN9 A0A2J7R4R0 A0A182YET2 D1FQ50 A0A182Q6H2 T1E347 A0A182NDY9 A0A182VPR0 A0A1B6K417 V5GW68 D6WSX9 A0A3L8D6R6 A0A067R407 A0A1B6IK82 A0A0A9XJP8 A0A026WRA3 A0A146KW89 A0A182LXT4 A0A182REB0 A0A182JXH1 A0A182J482 T1J6P0 A0A1B0GQC8 A0A0K8TQT7 A0A1B6KKT7 A0A146M9C8 B4JI06 A0A1B6MGM3 B0WNE9 A0A2H1W5U6 A0A232EX76 K7J3X9 A0A195BJ79 A0A158NA73 B4K4I0 N6UHM1 A0A2R7VZE8 A0A195FRC5 A0A0P4WD74 A0A1L8E016 A0A1L8EF44 R4WIQ2 A0A1L8EF88 B4LVY8 A0A1L8E0A0 A0A336L460 W8BWU4
Pubmed
26227816
26354079
28756777
22118469
20798317
17510324
+ More
19178717 20920257 23761445 24330624 27129103 20966253 12364791 14747013 17210077 29403074 17244540 24945155 26483478 21282665 21719571 26334808 25244985 18362917 19820115 30249741 24845553 25401762 24508170 26823975 26369729 17994087 28648823 20075255 21347285 23537049 23691247 24495485
19178717 20920257 23761445 24330624 27129103 20966253 12364791 14747013 17210077 29403074 17244540 24945155 26483478 21282665 21719571 26334808 25244985 18362917 19820115 30249741 24845553 25401762 24508170 26823975 26369729 17994087 28648823 20075255 21347285 23537049 23691247 24495485
EMBL
DQ443376
ABF51465.1
JTDY01007883
KOB64888.1
KQ459604
KPI92118.1
+ More
KQ460205 KPJ17040.1 GBRD01010709 JAG55115.1 KZ149945 PZC76810.1 RSAL01000004 RVE54546.1 AGBW02014918 OWR40883.1 NWSH01001499 PCG71054.1 PCG71055.1 ODYU01006087 SOQ47623.1 HM536614 ADO95153.1 KZ150148 PZC72864.1 NWSH01002103 PCG69177.1 CVRI01000039 CRK94684.1 FJ458409 ACL37991.1 GL439339 EFN67415.1 CH477253 EAT45962.1 EU934325 ADMH02001905 ACI30103.1 ETN60559.1 GANO01002412 JAB57459.1 LBMM01000967 KMQ97137.1 KQ982446 KYQ56327.1 GALA01000155 JAA94697.1 KQ978009 KYM98130.1 GAMD01003311 JAA98279.1 GGFK01004271 MBW37592.1 GFTR01001745 JAW14681.1 LJIG01016396 KRT80728.1 KQ980734 KYN13842.1 CAQQ02077811 CAQQ02077812 CAQQ02077813 GEDC01029991 JAS07307.1 GGFM01001126 MBW21877.1 GGFJ01010972 MBW60113.1 GDKW01002385 JAI54210.1 ACPB03001069 GAHY01002104 JAA75406.1 AAAB01008952 EAA10598.2 APCN01005975 PYGN01000288 PSN49237.1 JXUM01060179 AY826161 GAPW01006356 GAPW01006355 GAPW01006354 KQ562091 AAV90733.1 JAC07242.1 KXJ76715.1 GALA01000497 JAA94355.1 GL766762 EFZ14637.1 GECL01003515 JAP02609.1 GL888529 EGI59696.1 GECZ01007667 JAS62102.1 GBGD01003569 JAC85320.1 NEVH01007402 PNF35819.1 EZ419950 ACZ28305.1 AXCN02002212 GALA01000154 JAA94698.1 GECU01001505 JAT06202.1 GALX01002544 JAB65922.1 KQ971352 EFA07467.2 QOIP01000012 RLU15881.1 KK852718 KDR17794.1 GECU01020380 JAS87326.1 GBHO01022657 GBHO01022652 GBHO01011467 JAG20947.1 JAG20952.1 JAG32137.1 KK107131 EZA58201.1 GDHC01018111 JAQ00518.1 AXCM01000127 JH431880 AJVK01016284 GDAI01001333 JAI16270.1 GEBQ01027905 JAT12072.1 GDHC01002281 JAQ16348.1 CH916369 EDV93926.1 GEBQ01004946 JAT35031.1 DS232010 EDS31603.1 ODYU01006502 SOQ48393.1 NNAY01001764 OXU22986.1 AAZX01000988 KQ976455 KYM85156.1 ADTU01010056 ADTU01010057 CH933806 EDW13932.1 APGK01034997 KB740923 ENN78132.1 KK854191 PTY12867.1 KQ981305 KYN42981.1 GDRN01079695 GDRN01079693 JAI62341.1 GFDF01001986 JAV12098.1 GFDG01001452 JAV17347.1 AK417340 BAN20555.1 GFDG01001453 JAV17346.1 CH940650 EDW66493.2 GFDF01001985 JAV12099.1 UFQS01000993 UFQT01000993 SSX08314.1 SSX28337.1 GAMC01000775 JAC05781.1
KQ460205 KPJ17040.1 GBRD01010709 JAG55115.1 KZ149945 PZC76810.1 RSAL01000004 RVE54546.1 AGBW02014918 OWR40883.1 NWSH01001499 PCG71054.1 PCG71055.1 ODYU01006087 SOQ47623.1 HM536614 ADO95153.1 KZ150148 PZC72864.1 NWSH01002103 PCG69177.1 CVRI01000039 CRK94684.1 FJ458409 ACL37991.1 GL439339 EFN67415.1 CH477253 EAT45962.1 EU934325 ADMH02001905 ACI30103.1 ETN60559.1 GANO01002412 JAB57459.1 LBMM01000967 KMQ97137.1 KQ982446 KYQ56327.1 GALA01000155 JAA94697.1 KQ978009 KYM98130.1 GAMD01003311 JAA98279.1 GGFK01004271 MBW37592.1 GFTR01001745 JAW14681.1 LJIG01016396 KRT80728.1 KQ980734 KYN13842.1 CAQQ02077811 CAQQ02077812 CAQQ02077813 GEDC01029991 JAS07307.1 GGFM01001126 MBW21877.1 GGFJ01010972 MBW60113.1 GDKW01002385 JAI54210.1 ACPB03001069 GAHY01002104 JAA75406.1 AAAB01008952 EAA10598.2 APCN01005975 PYGN01000288 PSN49237.1 JXUM01060179 AY826161 GAPW01006356 GAPW01006355 GAPW01006354 KQ562091 AAV90733.1 JAC07242.1 KXJ76715.1 GALA01000497 JAA94355.1 GL766762 EFZ14637.1 GECL01003515 JAP02609.1 GL888529 EGI59696.1 GECZ01007667 JAS62102.1 GBGD01003569 JAC85320.1 NEVH01007402 PNF35819.1 EZ419950 ACZ28305.1 AXCN02002212 GALA01000154 JAA94698.1 GECU01001505 JAT06202.1 GALX01002544 JAB65922.1 KQ971352 EFA07467.2 QOIP01000012 RLU15881.1 KK852718 KDR17794.1 GECU01020380 JAS87326.1 GBHO01022657 GBHO01022652 GBHO01011467 JAG20947.1 JAG20952.1 JAG32137.1 KK107131 EZA58201.1 GDHC01018111 JAQ00518.1 AXCM01000127 JH431880 AJVK01016284 GDAI01001333 JAI16270.1 GEBQ01027905 JAT12072.1 GDHC01002281 JAQ16348.1 CH916369 EDV93926.1 GEBQ01004946 JAT35031.1 DS232010 EDS31603.1 ODYU01006502 SOQ48393.1 NNAY01001764 OXU22986.1 AAZX01000988 KQ976455 KYM85156.1 ADTU01010056 ADTU01010057 CH933806 EDW13932.1 APGK01034997 KB740923 ENN78132.1 KK854191 PTY12867.1 KQ981305 KYN42981.1 GDRN01079695 GDRN01079693 JAI62341.1 GFDF01001986 JAV12098.1 GFDG01001452 JAV17347.1 AK417340 BAN20555.1 GFDG01001453 JAV17346.1 CH940650 EDW66493.2 GFDF01001985 JAV12099.1 UFQS01000993 UFQT01000993 SSX08314.1 SSX28337.1 GAMC01000775 JAC05781.1
Proteomes
UP000037510
UP000053268
UP000053240
UP000283053
UP000007151
UP000218220
+ More
UP000183832 UP000000311 UP000008820 UP000000673 UP000075885 UP000036403 UP000075809 UP000078542 UP000078492 UP000015102 UP000076407 UP000069272 UP000015103 UP000075882 UP000007062 UP000075840 UP000245037 UP000069940 UP000249989 UP000007755 UP000075902 UP000075903 UP000192223 UP000235965 UP000076408 UP000075886 UP000075884 UP000075920 UP000007266 UP000279307 UP000027135 UP000053097 UP000075883 UP000075900 UP000075881 UP000075880 UP000092462 UP000001070 UP000002320 UP000215335 UP000002358 UP000078540 UP000005205 UP000009192 UP000019118 UP000078541 UP000008792
UP000183832 UP000000311 UP000008820 UP000000673 UP000075885 UP000036403 UP000075809 UP000078542 UP000078492 UP000015102 UP000076407 UP000069272 UP000015103 UP000075882 UP000007062 UP000075840 UP000245037 UP000069940 UP000249989 UP000007755 UP000075902 UP000075903 UP000192223 UP000235965 UP000076408 UP000075886 UP000075884 UP000075920 UP000007266 UP000279307 UP000027135 UP000053097 UP000075883 UP000075900 UP000075881 UP000075880 UP000092462 UP000001070 UP000002320 UP000215335 UP000002358 UP000078540 UP000005205 UP000009192 UP000019118 UP000078541 UP000008792
PRIDE
Interpro
Gene 3D
ProteinModelPortal
Q1HPM6
A0A0L7KNK4
A0A194PGG3
A0A194RI86
A0A0K8SQL0
A0A2W1BVJ1
+ More
A0A3S2LVB0 A0A212EHB4 A0A2A4JHJ2 A0A2H1W3L0 E3UPD0 A0A2W1BGQ5 A0A2A4JAN6 A0A1J1I514 B8XY08 E2AGN2 Q17H51 A0A1S4F2T8 B6DDV7 A0A182PSQ5 U5EUA2 A0A0J7L2W9 A0A151X7I3 T1E3Q7 A0A195CBQ6 T1DEZ9 A0A2M4AA23 A0A224Y2F2 A0A0T6B055 A0A195DLY5 T1GY90 A0A182XDQ5 A0A1B6C1C8 A0A2M3Z030 A0A182FP58 A0A2M4C473 A0A0P4VHT1 R4FLH7 A0A182LNN7 Q7Q7P4 A0A182IDG4 A0A2P8YYB5 Q5MIP9 T1DFP1 E9IXS2 A0A0V0G3G2 F4X1G8 A0A1B6GI34 A0A069DNC6 A0A182TQ12 A0A182V2I8 A0A1W4XIN9 A0A2J7R4R0 A0A182YET2 D1FQ50 A0A182Q6H2 T1E347 A0A182NDY9 A0A182VPR0 A0A1B6K417 V5GW68 D6WSX9 A0A3L8D6R6 A0A067R407 A0A1B6IK82 A0A0A9XJP8 A0A026WRA3 A0A146KW89 A0A182LXT4 A0A182REB0 A0A182JXH1 A0A182J482 T1J6P0 A0A1B0GQC8 A0A0K8TQT7 A0A1B6KKT7 A0A146M9C8 B4JI06 A0A1B6MGM3 B0WNE9 A0A2H1W5U6 A0A232EX76 K7J3X9 A0A195BJ79 A0A158NA73 B4K4I0 N6UHM1 A0A2R7VZE8 A0A195FRC5 A0A0P4WD74 A0A1L8E016 A0A1L8EF44 R4WIQ2 A0A1L8EF88 B4LVY8 A0A1L8E0A0 A0A336L460 W8BWU4
A0A3S2LVB0 A0A212EHB4 A0A2A4JHJ2 A0A2H1W3L0 E3UPD0 A0A2W1BGQ5 A0A2A4JAN6 A0A1J1I514 B8XY08 E2AGN2 Q17H51 A0A1S4F2T8 B6DDV7 A0A182PSQ5 U5EUA2 A0A0J7L2W9 A0A151X7I3 T1E3Q7 A0A195CBQ6 T1DEZ9 A0A2M4AA23 A0A224Y2F2 A0A0T6B055 A0A195DLY5 T1GY90 A0A182XDQ5 A0A1B6C1C8 A0A2M3Z030 A0A182FP58 A0A2M4C473 A0A0P4VHT1 R4FLH7 A0A182LNN7 Q7Q7P4 A0A182IDG4 A0A2P8YYB5 Q5MIP9 T1DFP1 E9IXS2 A0A0V0G3G2 F4X1G8 A0A1B6GI34 A0A069DNC6 A0A182TQ12 A0A182V2I8 A0A1W4XIN9 A0A2J7R4R0 A0A182YET2 D1FQ50 A0A182Q6H2 T1E347 A0A182NDY9 A0A182VPR0 A0A1B6K417 V5GW68 D6WSX9 A0A3L8D6R6 A0A067R407 A0A1B6IK82 A0A0A9XJP8 A0A026WRA3 A0A146KW89 A0A182LXT4 A0A182REB0 A0A182JXH1 A0A182J482 T1J6P0 A0A1B0GQC8 A0A0K8TQT7 A0A1B6KKT7 A0A146M9C8 B4JI06 A0A1B6MGM3 B0WNE9 A0A2H1W5U6 A0A232EX76 K7J3X9 A0A195BJ79 A0A158NA73 B4K4I0 N6UHM1 A0A2R7VZE8 A0A195FRC5 A0A0P4WD74 A0A1L8E016 A0A1L8EF44 R4WIQ2 A0A1L8EF88 B4LVY8 A0A1L8E0A0 A0A336L460 W8BWU4
PDB
5FIL
E-value=9.82846e-06,
Score=110
Ontologies
PATHWAY
GO
PANTHER
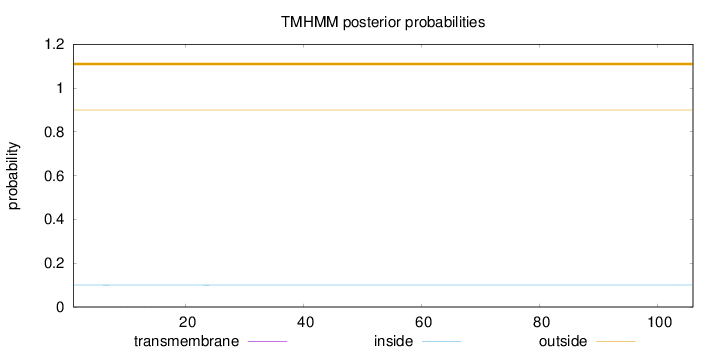
Topology
Subcellular location
Mitochondrion
Mitochondrion inner membrane
Mitochondrion inner membrane
Length:
106
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00752
Exp number, first 60 AAs:
0.00752
Total prob of N-in:
0.10031
outside
1 - 106
Population Genetic Test Statistics
Pi
405.393732
Theta
232.584468
Tajima's D
2.316951
CLR
0.172716
CSRT
0.926303684815759
Interpretation
Uncertain
