Gene
KWMTBOMO14937 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA000803
Annotation
PREDICTED:_viral_IAP-associated_factor_homolog_[Amyelois_transitella]
Full name
Viral IAP-associated factor homolog
Location in the cell
Cytoplasmic Reliability : 3.097
Sequence
CDS
ATGCAGAACCCCAATGAAGATACAGAATGGAACGATACCCTTCGAAAGATGGGTATTATACCGCAGAAAGAAGCGGAGTTTTCGGAGGCCGATATTGTGAACATGATAGAGGACACAATACAGCAAAAGGAAGCTGAAAAACAAAAACAATTATCTGAGTTAGACCTTGATGGATTGGACGAACTTGAAGATTCAGAAGATGAGGCTGTTATAGAAGAGTACCGCAGGAAACGATTAGCAGAAATTAAGAAGCTGTCTGAGAAGCCTAGATTCGGAGAAGTGCGAGAAGTCTCTGGTCAAGATTATGTGCAGGAGGTTAACAAGGCTGGTCAAGACATTTGGGTGGTCATTCATTTGTACAAACAGGGAATACAACAATGTGCACTAATAAATCAACATCTACGAGAATTAGCAGTCAAGTTTCCCTACACAAAATTCTTGAAAGCCATTGCTCAGACTTGCATCCCAAACTTCCCAGAGAGGAATCTGCCTAGTTTATTTGTTTACTTTGAGGGCGAAATGAAGCAGCAGTTTGTGGGGTCCCACGAACTAAGGGGCACCAGTTTGACTTGTGATGAACTCGAATACATACTCGGTAAGGTTGGAGCTGTCGAAACGAGCATAACAGAAGATCCCCGACCAAAGATCAAAGACAAGTTGTTTACTGACCTTGGAAGTAATGACTGGTGA
Protein
MQNPNEDTEWNDTLRKMGIIPQKEAEFSEADIVNMIEDTIQQKEAEKQKQLSELDLDGLDELEDSEDEAVIEEYRRKRLAEIKKLSEKPRFGEVREVSGQDYVQEVNKAGQDIWVVIHLYKQGIQQCALINQHLRELAVKFPYTKFLKAIAQTCIPNFPERNLPSLFVYFEGEMKQQFVGSHELRGTSLTCDELEYILGKVGAVETSITEDPRPKIKDKLFTDLGSNDW
Summary
Description
Modulates the activation of caspases during apoptosis.
Similarity
Belongs to the phosducin family.
Keywords
Apoptosis
Complete proteome
Cytoplasm
Phosphoprotein
Reference proteome
Feature
chain Viral IAP-associated factor homolog
Uniprot
H9IU73
A0A212EHD1
S4PET9
A0A2A4JH64
A0A2W1BUD1
A0A2H1X233
+ More
A0A1E1WES2 I4DJF5 A0A3S2LD36 A0A194RHX9 A0A067RI91 V5FV39 A0A2J7R8K6 D6WHT2 J3JV15 A0A336LYM8 E0VA02 A0A1W4X768 Q0IEX9 A0A1S4FHK5 A0A1Y1K4S2 A0A1B0CKW1 B0WLG3 A0A023EKG8 A0A1L8DMP4 U5ETY0 A0A1B0CZ25 A0A1J1II09 A0A2M3ZC53 A0A182FBW0 A0A2M4BYM2 N6U3J8 B4L015 A0A2M4ASG6 A0A1B6EJE2 A0A2M4BYL5 W5JEP4 B4LHT9 U4UL02 A0A1Q3FC55 A0A3B0JUF9 A0A0A9VXL7 B4GQY2 Q7PGA7 Q2M0Y3 A0A0L0BVU9 K7J2J4 A0A182IJW7 B4MKX1 A0A182PLT9 A0A182SIA4 B3NH48 A0A224XVH5 A0A1B6KM10 A0A182U4L0 A0A182RJT3 W8BCQ0 A0A182JV25 B4J0Q0 Q8MR62 A0A182M3U0 A0A0P4VTA1 A0A1L8EDP1 B4PG77 A0A182QPQ0 B3M4N8 R4G4M3 A0A182HXE2 A0A1B6LH43 A0A2R7VTP2 A0A034WN27 A0A1W4VV88 A0A087ZY37 B4HFH3 B4QQR8 A0A182VL48 A0A0V0G6D5 A0A182X1N1 A0A069DPZ0 A0A0K8VLM0 A0A182YIU1 A0A182NDF3 A0A2A3EPU8 A0A182W3X5 A0A023F8D7 A0A1I8PBL9 A0A1D2NCC8 A0A026VT20 A0A0K8TY23 A0A2R5LIJ4 A0A0M5J0T8 A0A1A9UQQ6 A0A158NRK6 F4WBQ5 A0A170XJC1 A0A023GGI3 A0A1A9XYW1 T1P8F8 E9IG18 A0A1I8MRA0
A0A1E1WES2 I4DJF5 A0A3S2LD36 A0A194RHX9 A0A067RI91 V5FV39 A0A2J7R8K6 D6WHT2 J3JV15 A0A336LYM8 E0VA02 A0A1W4X768 Q0IEX9 A0A1S4FHK5 A0A1Y1K4S2 A0A1B0CKW1 B0WLG3 A0A023EKG8 A0A1L8DMP4 U5ETY0 A0A1B0CZ25 A0A1J1II09 A0A2M3ZC53 A0A182FBW0 A0A2M4BYM2 N6U3J8 B4L015 A0A2M4ASG6 A0A1B6EJE2 A0A2M4BYL5 W5JEP4 B4LHT9 U4UL02 A0A1Q3FC55 A0A3B0JUF9 A0A0A9VXL7 B4GQY2 Q7PGA7 Q2M0Y3 A0A0L0BVU9 K7J2J4 A0A182IJW7 B4MKX1 A0A182PLT9 A0A182SIA4 B3NH48 A0A224XVH5 A0A1B6KM10 A0A182U4L0 A0A182RJT3 W8BCQ0 A0A182JV25 B4J0Q0 Q8MR62 A0A182M3U0 A0A0P4VTA1 A0A1L8EDP1 B4PG77 A0A182QPQ0 B3M4N8 R4G4M3 A0A182HXE2 A0A1B6LH43 A0A2R7VTP2 A0A034WN27 A0A1W4VV88 A0A087ZY37 B4HFH3 B4QQR8 A0A182VL48 A0A0V0G6D5 A0A182X1N1 A0A069DPZ0 A0A0K8VLM0 A0A182YIU1 A0A182NDF3 A0A2A3EPU8 A0A182W3X5 A0A023F8D7 A0A1I8PBL9 A0A1D2NCC8 A0A026VT20 A0A0K8TY23 A0A2R5LIJ4 A0A0M5J0T8 A0A1A9UQQ6 A0A158NRK6 F4WBQ5 A0A170XJC1 A0A023GGI3 A0A1A9XYW1 T1P8F8 E9IG18 A0A1I8MRA0
Pubmed
19121390
22118469
23622113
28756777
22651552
26354079
+ More
24845553 18362917 19820115 22516182 20566863 17510324 28004739 24945155 26483478 23537049 17994087 20920257 23761445 25401762 26823975 12364791 14747013 17210077 15632085 26108605 20075255 24495485 15371430 10731132 12537572 12537569 18327897 27129103 17550304 25348373 22936249 26334808 25244985 25474469 27289101 24508170 21347285 21719571 21282665 25315136
24845553 18362917 19820115 22516182 20566863 17510324 28004739 24945155 26483478 23537049 17994087 20920257 23761445 25401762 26823975 12364791 14747013 17210077 15632085 26108605 20075255 24495485 15371430 10731132 12537572 12537569 18327897 27129103 17550304 25348373 22936249 26334808 25244985 25474469 27289101 24508170 21347285 21719571 21282665 25315136
EMBL
BABH01034205
AGBW02014918
OWR40880.1
GAIX01004342
JAA88218.1
NWSH01001499
+ More
PCG71058.1 KZ149945 PZC76807.1 ODYU01012810 SOQ59302.1 GDQN01005598 JAT85456.1 AK401423 KQ459604 BAM18045.1 KPI92120.1 RSAL01000038 RVE51067.1 KQ460205 KPJ17044.1 KK852618 KDR20134.1 GALX01006956 JAB61510.1 NEVH01006721 PNF37168.1 KQ971321 EFA00689.1 BT127081 AEE62043.1 UFQT01000292 SSX22910.1 DS235002 EEB10208.1 CH477461 EAT40562.1 GEZM01092789 JAV56462.1 AJWK01016666 DS231984 EDS30456.1 JXUM01144899 GAPW01003940 KQ569880 JAC09658.1 KXJ68494.1 GFDF01006450 JAV07634.1 GANO01001716 JAB58155.1 AJVK01009459 CVRI01000054 CRK99885.1 GGFM01005289 MBW26040.1 GGFJ01009009 MBW58150.1 APGK01050864 KB741178 ENN73142.1 CH933809 EDW19050.1 GGFK01010217 MBW43538.1 GECZ01031743 GECZ01019919 GECZ01014555 GECZ01007915 JAS38026.1 JAS49850.1 JAS55214.1 JAS61854.1 GGFJ01009008 MBW58149.1 ADMH02001609 ETN61813.1 CH940647 EDW70664.1 KB632256 ERL90690.1 GFDL01009977 JAV25068.1 OUUW01000009 SPP84693.1 GBHO01044666 GBRD01014413 GBRD01011352 GDHC01011819 GDHC01006484 JAF98937.1 JAG51413.1 JAQ06810.1 JAQ12145.1 CH479188 EDW40167.1 AAAB01008849 EAA45004.3 CH379069 EAL30793.1 JRES01001255 KNC24133.1 CH963847 EDW72896.1 CH954178 EDV51505.1 GFTR01003914 JAW12512.1 GEBQ01027723 JAT12254.1 GAMC01011752 JAB94803.1 CH916366 EDV97905.1 AF110513 AE014296 AY122105 AAF49974.2 AAG21889.1 AAM52617.1 AXCM01000614 GDKW01002418 JAI54177.1 GFDG01001959 JAV16840.1 CM000159 EDW94241.1 AXCN02002125 CH902618 EDV39437.1 ACPB03003996 GAHY01000651 JAA76859.1 APCN01005189 GEBQ01017019 GEBQ01006204 JAT22958.1 JAT33773.1 KK854061 PTY10508.1 GAKP01003240 JAC55712.1 CH480815 EDW41204.1 CM000363 CM002912 EDX10170.1 KMY99130.1 GECL01002574 JAP03550.1 GBGD01002746 JAC86143.1 GDHF01028484 GDHF01012571 JAI23830.1 JAI39743.1 KZ288197 PBC33748.1 GBBI01001152 JAC17560.1 LJIJ01000092 ODN02913.1 KK108116 EZA46887.1 GDHF01032945 GDHF01001561 GDHF01001006 JAI19369.1 JAI50753.1 JAI51308.1 GGLE01005119 MBY09245.1 CP012525 ALC44374.1 ADTU01023988 GL888066 EGI68333.1 GEMB01004350 JAR98923.1 GBBM01002327 JAC33091.1 KA644929 AFP59558.1 GL762910 EFZ20449.1
PCG71058.1 KZ149945 PZC76807.1 ODYU01012810 SOQ59302.1 GDQN01005598 JAT85456.1 AK401423 KQ459604 BAM18045.1 KPI92120.1 RSAL01000038 RVE51067.1 KQ460205 KPJ17044.1 KK852618 KDR20134.1 GALX01006956 JAB61510.1 NEVH01006721 PNF37168.1 KQ971321 EFA00689.1 BT127081 AEE62043.1 UFQT01000292 SSX22910.1 DS235002 EEB10208.1 CH477461 EAT40562.1 GEZM01092789 JAV56462.1 AJWK01016666 DS231984 EDS30456.1 JXUM01144899 GAPW01003940 KQ569880 JAC09658.1 KXJ68494.1 GFDF01006450 JAV07634.1 GANO01001716 JAB58155.1 AJVK01009459 CVRI01000054 CRK99885.1 GGFM01005289 MBW26040.1 GGFJ01009009 MBW58150.1 APGK01050864 KB741178 ENN73142.1 CH933809 EDW19050.1 GGFK01010217 MBW43538.1 GECZ01031743 GECZ01019919 GECZ01014555 GECZ01007915 JAS38026.1 JAS49850.1 JAS55214.1 JAS61854.1 GGFJ01009008 MBW58149.1 ADMH02001609 ETN61813.1 CH940647 EDW70664.1 KB632256 ERL90690.1 GFDL01009977 JAV25068.1 OUUW01000009 SPP84693.1 GBHO01044666 GBRD01014413 GBRD01011352 GDHC01011819 GDHC01006484 JAF98937.1 JAG51413.1 JAQ06810.1 JAQ12145.1 CH479188 EDW40167.1 AAAB01008849 EAA45004.3 CH379069 EAL30793.1 JRES01001255 KNC24133.1 CH963847 EDW72896.1 CH954178 EDV51505.1 GFTR01003914 JAW12512.1 GEBQ01027723 JAT12254.1 GAMC01011752 JAB94803.1 CH916366 EDV97905.1 AF110513 AE014296 AY122105 AAF49974.2 AAG21889.1 AAM52617.1 AXCM01000614 GDKW01002418 JAI54177.1 GFDG01001959 JAV16840.1 CM000159 EDW94241.1 AXCN02002125 CH902618 EDV39437.1 ACPB03003996 GAHY01000651 JAA76859.1 APCN01005189 GEBQ01017019 GEBQ01006204 JAT22958.1 JAT33773.1 KK854061 PTY10508.1 GAKP01003240 JAC55712.1 CH480815 EDW41204.1 CM000363 CM002912 EDX10170.1 KMY99130.1 GECL01002574 JAP03550.1 GBGD01002746 JAC86143.1 GDHF01028484 GDHF01012571 JAI23830.1 JAI39743.1 KZ288197 PBC33748.1 GBBI01001152 JAC17560.1 LJIJ01000092 ODN02913.1 KK108116 EZA46887.1 GDHF01032945 GDHF01001561 GDHF01001006 JAI19369.1 JAI50753.1 JAI51308.1 GGLE01005119 MBY09245.1 CP012525 ALC44374.1 ADTU01023988 GL888066 EGI68333.1 GEMB01004350 JAR98923.1 GBBM01002327 JAC33091.1 KA644929 AFP59558.1 GL762910 EFZ20449.1
Proteomes
UP000005204
UP000007151
UP000218220
UP000053268
UP000283053
UP000053240
+ More
UP000027135 UP000235965 UP000007266 UP000009046 UP000192223 UP000008820 UP000092461 UP000002320 UP000069940 UP000249989 UP000092462 UP000183832 UP000069272 UP000019118 UP000009192 UP000000673 UP000008792 UP000030742 UP000268350 UP000008744 UP000007062 UP000001819 UP000037069 UP000002358 UP000075880 UP000007798 UP000075885 UP000075901 UP000008711 UP000075902 UP000075900 UP000075881 UP000001070 UP000000803 UP000075883 UP000002282 UP000075886 UP000007801 UP000015103 UP000075840 UP000192221 UP000005203 UP000001292 UP000000304 UP000075903 UP000076407 UP000076408 UP000075884 UP000242457 UP000075920 UP000095300 UP000094527 UP000053097 UP000092553 UP000078200 UP000005205 UP000007755 UP000092443 UP000095301
UP000027135 UP000235965 UP000007266 UP000009046 UP000192223 UP000008820 UP000092461 UP000002320 UP000069940 UP000249989 UP000092462 UP000183832 UP000069272 UP000019118 UP000009192 UP000000673 UP000008792 UP000030742 UP000268350 UP000008744 UP000007062 UP000001819 UP000037069 UP000002358 UP000075880 UP000007798 UP000075885 UP000075901 UP000008711 UP000075902 UP000075900 UP000075881 UP000001070 UP000000803 UP000075883 UP000002282 UP000075886 UP000007801 UP000015103 UP000075840 UP000192221 UP000005203 UP000001292 UP000000304 UP000075903 UP000076407 UP000076408 UP000075884 UP000242457 UP000075920 UP000095300 UP000094527 UP000053097 UP000092553 UP000078200 UP000005205 UP000007755 UP000092443 UP000095301
Pfam
PF02114 Phosducin
SUPFAM
SSF52833
SSF52833
ProteinModelPortal
H9IU73
A0A212EHD1
S4PET9
A0A2A4JH64
A0A2W1BUD1
A0A2H1X233
+ More
A0A1E1WES2 I4DJF5 A0A3S2LD36 A0A194RHX9 A0A067RI91 V5FV39 A0A2J7R8K6 D6WHT2 J3JV15 A0A336LYM8 E0VA02 A0A1W4X768 Q0IEX9 A0A1S4FHK5 A0A1Y1K4S2 A0A1B0CKW1 B0WLG3 A0A023EKG8 A0A1L8DMP4 U5ETY0 A0A1B0CZ25 A0A1J1II09 A0A2M3ZC53 A0A182FBW0 A0A2M4BYM2 N6U3J8 B4L015 A0A2M4ASG6 A0A1B6EJE2 A0A2M4BYL5 W5JEP4 B4LHT9 U4UL02 A0A1Q3FC55 A0A3B0JUF9 A0A0A9VXL7 B4GQY2 Q7PGA7 Q2M0Y3 A0A0L0BVU9 K7J2J4 A0A182IJW7 B4MKX1 A0A182PLT9 A0A182SIA4 B3NH48 A0A224XVH5 A0A1B6KM10 A0A182U4L0 A0A182RJT3 W8BCQ0 A0A182JV25 B4J0Q0 Q8MR62 A0A182M3U0 A0A0P4VTA1 A0A1L8EDP1 B4PG77 A0A182QPQ0 B3M4N8 R4G4M3 A0A182HXE2 A0A1B6LH43 A0A2R7VTP2 A0A034WN27 A0A1W4VV88 A0A087ZY37 B4HFH3 B4QQR8 A0A182VL48 A0A0V0G6D5 A0A182X1N1 A0A069DPZ0 A0A0K8VLM0 A0A182YIU1 A0A182NDF3 A0A2A3EPU8 A0A182W3X5 A0A023F8D7 A0A1I8PBL9 A0A1D2NCC8 A0A026VT20 A0A0K8TY23 A0A2R5LIJ4 A0A0M5J0T8 A0A1A9UQQ6 A0A158NRK6 F4WBQ5 A0A170XJC1 A0A023GGI3 A0A1A9XYW1 T1P8F8 E9IG18 A0A1I8MRA0
A0A1E1WES2 I4DJF5 A0A3S2LD36 A0A194RHX9 A0A067RI91 V5FV39 A0A2J7R8K6 D6WHT2 J3JV15 A0A336LYM8 E0VA02 A0A1W4X768 Q0IEX9 A0A1S4FHK5 A0A1Y1K4S2 A0A1B0CKW1 B0WLG3 A0A023EKG8 A0A1L8DMP4 U5ETY0 A0A1B0CZ25 A0A1J1II09 A0A2M3ZC53 A0A182FBW0 A0A2M4BYM2 N6U3J8 B4L015 A0A2M4ASG6 A0A1B6EJE2 A0A2M4BYL5 W5JEP4 B4LHT9 U4UL02 A0A1Q3FC55 A0A3B0JUF9 A0A0A9VXL7 B4GQY2 Q7PGA7 Q2M0Y3 A0A0L0BVU9 K7J2J4 A0A182IJW7 B4MKX1 A0A182PLT9 A0A182SIA4 B3NH48 A0A224XVH5 A0A1B6KM10 A0A182U4L0 A0A182RJT3 W8BCQ0 A0A182JV25 B4J0Q0 Q8MR62 A0A182M3U0 A0A0P4VTA1 A0A1L8EDP1 B4PG77 A0A182QPQ0 B3M4N8 R4G4M3 A0A182HXE2 A0A1B6LH43 A0A2R7VTP2 A0A034WN27 A0A1W4VV88 A0A087ZY37 B4HFH3 B4QQR8 A0A182VL48 A0A0V0G6D5 A0A182X1N1 A0A069DPZ0 A0A0K8VLM0 A0A182YIU1 A0A182NDF3 A0A2A3EPU8 A0A182W3X5 A0A023F8D7 A0A1I8PBL9 A0A1D2NCC8 A0A026VT20 A0A0K8TY23 A0A2R5LIJ4 A0A0M5J0T8 A0A1A9UQQ6 A0A158NRK6 F4WBQ5 A0A170XJC1 A0A023GGI3 A0A1A9XYW1 T1P8F8 E9IG18 A0A1I8MRA0
PDB
2DBC
E-value=7.38295e-30,
Score=322
Ontologies
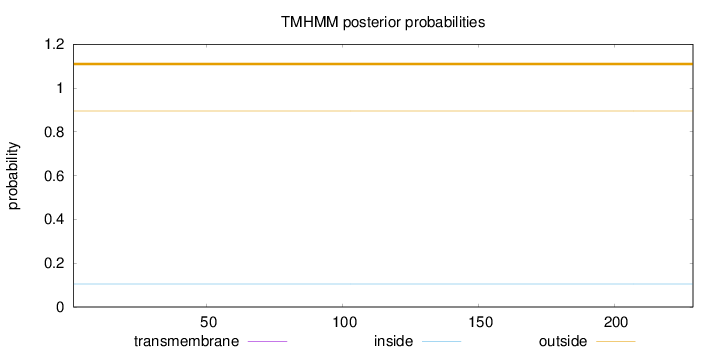
Topology
Subcellular location
Cytoplasm
Length:
229
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0004
Exp number, first 60 AAs:
0
Total prob of N-in:
0.10467
outside
1 - 229
Population Genetic Test Statistics
Pi
288.288218
Theta
216.14045
Tajima's D
0.87461
CLR
0.471307
CSRT
0.624268786560672
Interpretation
Possibly Positive selection
