Pre Gene Modal
BGIBMGA000767
Annotation
PREDICTED:_atlastin_isoform_X1_[Bombyx_mori]
Full name
Atlastin
Location in the cell
Cytoplasmic Reliability : 1.496
Sequence
CDS
ATGATGGCACATGTGACAGAGAAGCCGGCGCCGGCACCGCGTGGGCCGCACGGTGTCCAGGTCGTCGACACCGGCCCCGACCATACGTTCACCCTGGACGAGGAGGCCCTCAAGTCTCTGCTCCTGAAAGAAGATGTCAGAGACAGAGCCGTGGTCGTTATATCAGTTGCCGGAGCCTTCAGGAAAGGAAAGTCCTTCTTATTGGACTTCTTCCTGAGATATATGCACTATAAGTACGGCGCAGGCGGCGGCGGGAGCGAGTGGCTCGGTAGCGAGGAGGACCCCCTCCAGGGCTTCAGCTGGCGAGGGGGCTCCGAGAGGGACACGACCGGCATACTCATGTGGTCCGAGATATTCAAAGCGACGCTCGATGATGGTGAAAAGGTAGCGATAATATTGCTAGACACGCAAGGCGCCTTCGACAGTGAATCCACGGTGCGAGACTGCGCGACCGTGTTCGCTCTGTCCACGATGCTGTCGTCCGTTCAGATATACAACCTCTCGCAAAACATCCAGGAGGACGACCTCCAGCACCTTCAATTGTTCACAGAGTACGGTCGGCTGGCGCTCGAGGACGGGGGCCGGACTCCCTTCCAGCGGCTGCAGTTCCTCGTGCGCGACTGGAGCTTCCCCTACGAAGCGCCCTACGGAGCGGACGGCGGCATGCAGATACTGTCAAGAAGATTAAAGGTATCAGACAAACAGCATCCAGAGCTGCAGTCCCTCCGCAAGCACATAACGGCGTGCTTCTCGGAGCTGGCCTGCTTCCTCATGCCGCACCCGGGACTCAAAGTGGCCACCAACCCACAATTTGACGGCAGACTTGCAGATATCGAATCAGAGTTCAAACGCTCCCTCCAGCAGTTCGTGCCGATGCTGCTGGCTCCCGAGAACCTGGTGCCGAAGCTCATCAACGGTCAGAAGGTCAAGGCCAAGGACCTGGTCCAGTACTTCAAGTCGTACATGAACATCTACAAGGGCAACGAACTGCCCGAGCCCAAGAGCATGCTTGTGGCGACAGCGGAAGCCAACAACTTGACAGCTGTGGCGGAGGCCAAGGAGGTGTACACGCGGCTCATGGAGGAGGTGTGCGGGGGTGGGCGGCCCTACTTGCACGCCGACCTGCTGCTGACGGAGCACCAGCGCGTGCGGGACAAGGCCATGTACGCCTTCAACGCCAAGCGGAAGATGGGCGGCCGGGACTTCAGCCAGGCCTACTTCGACCAACTCGCACAGGACTTGGAGGAGCAGTTCCTGCAGTTCAAGGCGCACAACGAGAGCAAGAACATCTTCAAGGCGGCCCGGACGCCCTCCGTGTTCTTCGCGGTGGCGCTGTTCTTCTACGTGCTCAGCGGAGTGTTCGGGCTGCTGGGCCTCTACCCGCTCGCCAACGTCGCCAACTTCGCCATGGGCCTGGCCCTCCTCACGCTCGTGCTCTGGGCATACATCAGGTACAGCGGCGAGATGAGAGAGTTCGGCGTAACCATAGACGACACTGCGACGGTCATGTGGGATAATGTCCTGAAGCCGATGTACCAGACGTGCCTCGAGAAGGGCATGGAGCACGCGGCGGCGGCGGCCGTGGCGCCCCCCTCCCCCGCCCCCGGCGCCGCGCCCCCCGCCTTCGCGCACAACGGGAAGCACAAGCACTTGTGA
Protein
MMAHVTEKPAPAPRGPHGVQVVDTGPDHTFTLDEEALKSLLLKEDVRDRAVVVISVAGAFRKGKSFLLDFFLRYMHYKYGAGGGGSEWLGSEEDPLQGFSWRGGSERDTTGILMWSEIFKATLDDGEKVAIILLDTQGAFDSESTVRDCATVFALSTMLSSVQIYNLSQNIQEDDLQHLQLFTEYGRLALEDGGRTPFQRLQFLVRDWSFPYEAPYGADGGMQILSRRLKVSDKQHPELQSLRKHITACFSELACFLMPHPGLKVATNPQFDGRLADIESEFKRSLQQFVPMLLAPENLVPKLINGQKVKAKDLVQYFKSYMNIYKGNELPEPKSMLVATAEANNLTAVAEAKEVYTRLMEEVCGGGRPYLHADLLLTEHQRVRDKAMYAFNAKRKMGGRDFSQAYFDQLAQDLEEQFLQFKAHNESKNIFKAARTPSVFFAVALFFYVLSGVFGLLGLYPLANVANFAMGLALLTLVLWAYIRYSGEMREFGVTIDDTATVMWDNVLKPMYQTCLEKGMEHAAAAAVAPPSPAPGAAPPAFAHNGKHKHL
Summary
Description
GTPase tethering membranes through formation of trans-homooligomers and mediating homotypic fusion of endoplasmic reticulum membranes. Functions in endoplasmic reticulum tubular network biogenesis. May also regulate microtubule polymerization and Golgi biogenesis. Required for dopaminergic neurons survival and the growth of muscles and synapses at neuromuscular junctions.
Subunit
Homooligomer; trans-homooligomer between proteins on adjacent membranes. Interacts with spas; interaction may regulate microtubule dynamics.
Similarity
Belongs to the TRAFAC class dynamin-like GTPase superfamily. GB1/RHD3-type GTPase family. GB1 subfamily.
Keywords
3D-structure
Complete proteome
Endoplasmic reticulum
Golgi apparatus
GTP-binding
Hydrolase
Membrane
Nucleotide-binding
Phosphoprotein
Reference proteome
Transmembrane
Transmembrane helix
Feature
chain Atlastin
Uniprot
H9IU37
A0A1E1W9U5
A0A1E1VXT1
A0A2A4JE55
A0A2W1BT43
A0A182IZZ3
+ More
A0A182Q5E4 A0A182VZ50 Q17GF2 A0A182P550 A0A182N454 A0A182MAV6 A0A182K3G1 A0A182TUD6 Q7PUS9 A0A023EUV9 A0A084VG42 A0A1Y1KJA0 A0A182GUP1 A0A1Y1KFA2 A0A182R689 A0A2M3Z0Q2 A0A2M4A1J4 W5JH04 A0A2M4A1Q3 A0A2M4BJH5 T1DN87 A0A182FNI8 A0A182YAR7 A0A182X0D2 A0A182LR64 A0A1Q3G2Q7 A0A182I021 A0A0K8V1U1 A0A182VG90 W8BN84 B4JEK1 A0A034W9U4 A0A154P404 B3P712 A0A182T9R3 B4K505 B4PVR8 B4LW27 A0A1W4VPR3 B4HH45 B3M050 A0A0A1WFI4 A0A0B4LHV8 A0A0K8TP26 B4N8U3 Q9VC57 A0A1L8DFC2 Q29BY7 B4GPG3 A0A1L8DFN1 A0A3B0JFY6 I3RSF4 A0A0M4EDL5 K7IUU5 A0A0L0C6K7 E2AR16 D6WQT2 A0A1I8Q5V2 A0A1I8MAE9 A0A195BG18 F4X8E3 A0A195FDX7 E2B5K9 B0WND1 A0A1Q3FBH4 A0A151J9D5 A0A0C9QVF4 A0A151X9P7 A0A0C9QJW0 A0A195CBN9 A0A1B6BYG1 A0A1B6C1Y7 A0A2S2QTC8 A0A0A9XY37 A0A158NBV9 A0A026X1X4 A0A1A9ZZN1 V9IF45 A0A087ZZN6 A0A2A3ET24 D3TMN5 A0A1B0FBF0 A0A158NBW0 A0A146LV81 A0A212EHB8 U5EXR1 N6TK58 A0A0L7QTT4 A0A0M9A692 A0A067RT26 A0A1B6FS83 A0A1A9XTA1 A0A1A9V4Y1 A0A1B0CGH0
A0A182Q5E4 A0A182VZ50 Q17GF2 A0A182P550 A0A182N454 A0A182MAV6 A0A182K3G1 A0A182TUD6 Q7PUS9 A0A023EUV9 A0A084VG42 A0A1Y1KJA0 A0A182GUP1 A0A1Y1KFA2 A0A182R689 A0A2M3Z0Q2 A0A2M4A1J4 W5JH04 A0A2M4A1Q3 A0A2M4BJH5 T1DN87 A0A182FNI8 A0A182YAR7 A0A182X0D2 A0A182LR64 A0A1Q3G2Q7 A0A182I021 A0A0K8V1U1 A0A182VG90 W8BN84 B4JEK1 A0A034W9U4 A0A154P404 B3P712 A0A182T9R3 B4K505 B4PVR8 B4LW27 A0A1W4VPR3 B4HH45 B3M050 A0A0A1WFI4 A0A0B4LHV8 A0A0K8TP26 B4N8U3 Q9VC57 A0A1L8DFC2 Q29BY7 B4GPG3 A0A1L8DFN1 A0A3B0JFY6 I3RSF4 A0A0M4EDL5 K7IUU5 A0A0L0C6K7 E2AR16 D6WQT2 A0A1I8Q5V2 A0A1I8MAE9 A0A195BG18 F4X8E3 A0A195FDX7 E2B5K9 B0WND1 A0A1Q3FBH4 A0A151J9D5 A0A0C9QVF4 A0A151X9P7 A0A0C9QJW0 A0A195CBN9 A0A1B6BYG1 A0A1B6C1Y7 A0A2S2QTC8 A0A0A9XY37 A0A158NBV9 A0A026X1X4 A0A1A9ZZN1 V9IF45 A0A087ZZN6 A0A2A3ET24 D3TMN5 A0A1B0FBF0 A0A158NBW0 A0A146LV81 A0A212EHB8 U5EXR1 N6TK58 A0A0L7QTT4 A0A0M9A692 A0A067RT26 A0A1B6FS83 A0A1A9XTA1 A0A1A9V4Y1 A0A1B0CGH0
EC Number
3.6.5.-
Pubmed
19121390
28756777
17510324
12364791
14747013
17210077
+ More
24945155 24438588 28004739 26483478 20920257 23761445 25244985 20966253 24495485 17994087 25348373 18057021 17550304 25830018 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26369729 18327897 17030474 19341724 19633650 15632085 22328714 20075255 26108605 20798317 18362917 19820115 25315136 21719571 25401762 26823975 21347285 24508170 30249741 20353571 22118469 23537049 24845553
24945155 24438588 28004739 26483478 20920257 23761445 25244985 20966253 24495485 17994087 25348373 18057021 17550304 25830018 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26369729 18327897 17030474 19341724 19633650 15632085 22328714 20075255 26108605 20798317 18362917 19820115 25315136 21719571 25401762 26823975 21347285 24508170 30249741 20353571 22118469 23537049 24845553
EMBL
BABH01034204
GDQN01007282
GDQN01004744
JAT83772.1
JAT86310.1
GDQN01011573
+ More
GDQN01000546 JAT79481.1 JAT90508.1 NWSH01001828 PCG69988.1 KZ149945 PZC76805.1 AXCN02000025 CH477262 EAT45634.1 AXCM01017510 AAAB01008987 EAA01072.3 GAPW01000826 JAC12772.1 ATLV01012626 KE524806 KFB36936.1 GEZM01087174 JAV58867.1 JXUM01019135 JXUM01019136 JXUM01019137 JXUM01019138 JXUM01061069 JXUM01061070 KQ562133 KXJ76608.1 GEZM01087172 JAV58870.1 GGFM01001358 MBW22109.1 GGFK01001301 MBW34622.1 ADMH02001421 ETN62603.1 GGFK01001358 MBW34679.1 GGFJ01004064 MBW53205.1 GAMD01003100 JAA98490.1 GFDL01000940 JAV34105.1 APCN01002031 GDHF01022865 GDHF01019425 JAI29449.1 JAI32889.1 GAMC01015402 GAMC01015401 JAB91154.1 CH916369 EDV93132.1 GAKP01007876 GAKP01007875 JAC51077.1 KQ434809 KZC06646.1 CH954182 EDV53832.1 CH933806 EDW13976.2 KRG00752.1 CM000160 EDW98917.2 KRK04548.1 CH940650 EDW66532.1 KRF82754.1 CH480815 EDW43521.1 CH902617 EDV42007.2 KPU79506.1 GBXI01017553 GBXI01016851 GBXI01009293 JAC96738.1 JAC97440.1 JAD04999.1 AE014297 AHN57505.1 GDAI01001685 JAI15918.1 CH964232 EDW81544.2 KRF99479.1 KRF99480.1 AY069079 BT004893 GFDF01008925 JAV05159.1 CM000070 EAL26859.1 CH479186 EDW39046.1 GFDF01008914 JAV05170.1 OUUW01000005 SPP81304.1 JQ611724 AFK29239.1 CP012526 ALC45916.1 AAZX01002640 JRES01000835 KNC27900.1 GL441846 EFN64162.1 KQ971354 EFA07005.1 KQ976500 KYM83149.1 GL888932 EGI57148.1 KQ981673 KYN38407.1 GL445852 EFN89021.1 DS232010 EDS31585.1 GFDL01010176 JAV24869.1 KQ979433 KYN21563.1 GBYB01004652 JAG74419.1 KQ982373 KYQ57054.1 GBYB01003799 JAG73566.1 KQ978068 KYM97528.1 GEDC01031239 GEDC01019967 GEDC01002587 JAS06059.1 JAS17331.1 JAS34711.1 GEDC01030063 GEDC01024076 GEDC01009683 JAS07235.1 JAS13222.1 JAS27615.1 GGMS01011823 MBY81026.1 GBHO01021429 GBRD01002498 GDHC01012112 JAG22175.1 JAG63323.1 JAQ06517.1 ADTU01011332 ADTU01011333 ADTU01011334 KK107036 QOIP01000004 EZA62001.1 RLU23239.1 JR040781 AEY59292.1 KZ288191 PBC34452.1 EZ422687 ADD18963.1 CCAG010007921 GDHC01008359 JAQ10270.1 AGBW02014918 OWR40879.1 GANO01000820 JAB59051.1 APGK01035351 KB740923 KB632333 ENN78293.1 ERL92718.1 KQ414740 KOC62077.1 KQ435741 KOX76783.1 KK852433 KDR23965.1 GECZ01016693 JAS53076.1 AJWK01011095 AJWK01011096
GDQN01000546 JAT79481.1 JAT90508.1 NWSH01001828 PCG69988.1 KZ149945 PZC76805.1 AXCN02000025 CH477262 EAT45634.1 AXCM01017510 AAAB01008987 EAA01072.3 GAPW01000826 JAC12772.1 ATLV01012626 KE524806 KFB36936.1 GEZM01087174 JAV58867.1 JXUM01019135 JXUM01019136 JXUM01019137 JXUM01019138 JXUM01061069 JXUM01061070 KQ562133 KXJ76608.1 GEZM01087172 JAV58870.1 GGFM01001358 MBW22109.1 GGFK01001301 MBW34622.1 ADMH02001421 ETN62603.1 GGFK01001358 MBW34679.1 GGFJ01004064 MBW53205.1 GAMD01003100 JAA98490.1 GFDL01000940 JAV34105.1 APCN01002031 GDHF01022865 GDHF01019425 JAI29449.1 JAI32889.1 GAMC01015402 GAMC01015401 JAB91154.1 CH916369 EDV93132.1 GAKP01007876 GAKP01007875 JAC51077.1 KQ434809 KZC06646.1 CH954182 EDV53832.1 CH933806 EDW13976.2 KRG00752.1 CM000160 EDW98917.2 KRK04548.1 CH940650 EDW66532.1 KRF82754.1 CH480815 EDW43521.1 CH902617 EDV42007.2 KPU79506.1 GBXI01017553 GBXI01016851 GBXI01009293 JAC96738.1 JAC97440.1 JAD04999.1 AE014297 AHN57505.1 GDAI01001685 JAI15918.1 CH964232 EDW81544.2 KRF99479.1 KRF99480.1 AY069079 BT004893 GFDF01008925 JAV05159.1 CM000070 EAL26859.1 CH479186 EDW39046.1 GFDF01008914 JAV05170.1 OUUW01000005 SPP81304.1 JQ611724 AFK29239.1 CP012526 ALC45916.1 AAZX01002640 JRES01000835 KNC27900.1 GL441846 EFN64162.1 KQ971354 EFA07005.1 KQ976500 KYM83149.1 GL888932 EGI57148.1 KQ981673 KYN38407.1 GL445852 EFN89021.1 DS232010 EDS31585.1 GFDL01010176 JAV24869.1 KQ979433 KYN21563.1 GBYB01004652 JAG74419.1 KQ982373 KYQ57054.1 GBYB01003799 JAG73566.1 KQ978068 KYM97528.1 GEDC01031239 GEDC01019967 GEDC01002587 JAS06059.1 JAS17331.1 JAS34711.1 GEDC01030063 GEDC01024076 GEDC01009683 JAS07235.1 JAS13222.1 JAS27615.1 GGMS01011823 MBY81026.1 GBHO01021429 GBRD01002498 GDHC01012112 JAG22175.1 JAG63323.1 JAQ06517.1 ADTU01011332 ADTU01011333 ADTU01011334 KK107036 QOIP01000004 EZA62001.1 RLU23239.1 JR040781 AEY59292.1 KZ288191 PBC34452.1 EZ422687 ADD18963.1 CCAG010007921 GDHC01008359 JAQ10270.1 AGBW02014918 OWR40879.1 GANO01000820 JAB59051.1 APGK01035351 KB740923 KB632333 ENN78293.1 ERL92718.1 KQ414740 KOC62077.1 KQ435741 KOX76783.1 KK852433 KDR23965.1 GECZ01016693 JAS53076.1 AJWK01011095 AJWK01011096
Proteomes
UP000005204
UP000218220
UP000075880
UP000075886
UP000075920
UP000008820
+ More
UP000075885 UP000075884 UP000075883 UP000075881 UP000075902 UP000007062 UP000030765 UP000069940 UP000249989 UP000075900 UP000000673 UP000069272 UP000076408 UP000076407 UP000075882 UP000075840 UP000075903 UP000001070 UP000076502 UP000008711 UP000075901 UP000009192 UP000002282 UP000008792 UP000192221 UP000001292 UP000007801 UP000000803 UP000007798 UP000001819 UP000008744 UP000268350 UP000092553 UP000002358 UP000037069 UP000000311 UP000007266 UP000095300 UP000095301 UP000078540 UP000007755 UP000078541 UP000008237 UP000002320 UP000078492 UP000075809 UP000078542 UP000005205 UP000053097 UP000279307 UP000092445 UP000005203 UP000242457 UP000092444 UP000007151 UP000019118 UP000030742 UP000053825 UP000053105 UP000027135 UP000092443 UP000078200 UP000092461
UP000075885 UP000075884 UP000075883 UP000075881 UP000075902 UP000007062 UP000030765 UP000069940 UP000249989 UP000075900 UP000000673 UP000069272 UP000076408 UP000076407 UP000075882 UP000075840 UP000075903 UP000001070 UP000076502 UP000008711 UP000075901 UP000009192 UP000002282 UP000008792 UP000192221 UP000001292 UP000007801 UP000000803 UP000007798 UP000001819 UP000008744 UP000268350 UP000092553 UP000002358 UP000037069 UP000000311 UP000007266 UP000095300 UP000095301 UP000078540 UP000007755 UP000078541 UP000008237 UP000002320 UP000078492 UP000075809 UP000078542 UP000005205 UP000053097 UP000279307 UP000092445 UP000005203 UP000242457 UP000092444 UP000007151 UP000019118 UP000030742 UP000053825 UP000053105 UP000027135 UP000092443 UP000078200 UP000092461
Interpro
ProteinModelPortal
H9IU37
A0A1E1W9U5
A0A1E1VXT1
A0A2A4JE55
A0A2W1BT43
A0A182IZZ3
+ More
A0A182Q5E4 A0A182VZ50 Q17GF2 A0A182P550 A0A182N454 A0A182MAV6 A0A182K3G1 A0A182TUD6 Q7PUS9 A0A023EUV9 A0A084VG42 A0A1Y1KJA0 A0A182GUP1 A0A1Y1KFA2 A0A182R689 A0A2M3Z0Q2 A0A2M4A1J4 W5JH04 A0A2M4A1Q3 A0A2M4BJH5 T1DN87 A0A182FNI8 A0A182YAR7 A0A182X0D2 A0A182LR64 A0A1Q3G2Q7 A0A182I021 A0A0K8V1U1 A0A182VG90 W8BN84 B4JEK1 A0A034W9U4 A0A154P404 B3P712 A0A182T9R3 B4K505 B4PVR8 B4LW27 A0A1W4VPR3 B4HH45 B3M050 A0A0A1WFI4 A0A0B4LHV8 A0A0K8TP26 B4N8U3 Q9VC57 A0A1L8DFC2 Q29BY7 B4GPG3 A0A1L8DFN1 A0A3B0JFY6 I3RSF4 A0A0M4EDL5 K7IUU5 A0A0L0C6K7 E2AR16 D6WQT2 A0A1I8Q5V2 A0A1I8MAE9 A0A195BG18 F4X8E3 A0A195FDX7 E2B5K9 B0WND1 A0A1Q3FBH4 A0A151J9D5 A0A0C9QVF4 A0A151X9P7 A0A0C9QJW0 A0A195CBN9 A0A1B6BYG1 A0A1B6C1Y7 A0A2S2QTC8 A0A0A9XY37 A0A158NBV9 A0A026X1X4 A0A1A9ZZN1 V9IF45 A0A087ZZN6 A0A2A3ET24 D3TMN5 A0A1B0FBF0 A0A158NBW0 A0A146LV81 A0A212EHB8 U5EXR1 N6TK58 A0A0L7QTT4 A0A0M9A692 A0A067RT26 A0A1B6FS83 A0A1A9XTA1 A0A1A9V4Y1 A0A1B0CGH0
A0A182Q5E4 A0A182VZ50 Q17GF2 A0A182P550 A0A182N454 A0A182MAV6 A0A182K3G1 A0A182TUD6 Q7PUS9 A0A023EUV9 A0A084VG42 A0A1Y1KJA0 A0A182GUP1 A0A1Y1KFA2 A0A182R689 A0A2M3Z0Q2 A0A2M4A1J4 W5JH04 A0A2M4A1Q3 A0A2M4BJH5 T1DN87 A0A182FNI8 A0A182YAR7 A0A182X0D2 A0A182LR64 A0A1Q3G2Q7 A0A182I021 A0A0K8V1U1 A0A182VG90 W8BN84 B4JEK1 A0A034W9U4 A0A154P404 B3P712 A0A182T9R3 B4K505 B4PVR8 B4LW27 A0A1W4VPR3 B4HH45 B3M050 A0A0A1WFI4 A0A0B4LHV8 A0A0K8TP26 B4N8U3 Q9VC57 A0A1L8DFC2 Q29BY7 B4GPG3 A0A1L8DFN1 A0A3B0JFY6 I3RSF4 A0A0M4EDL5 K7IUU5 A0A0L0C6K7 E2AR16 D6WQT2 A0A1I8Q5V2 A0A1I8MAE9 A0A195BG18 F4X8E3 A0A195FDX7 E2B5K9 B0WND1 A0A1Q3FBH4 A0A151J9D5 A0A0C9QVF4 A0A151X9P7 A0A0C9QJW0 A0A195CBN9 A0A1B6BYG1 A0A1B6C1Y7 A0A2S2QTC8 A0A0A9XY37 A0A158NBV9 A0A026X1X4 A0A1A9ZZN1 V9IF45 A0A087ZZN6 A0A2A3ET24 D3TMN5 A0A1B0FBF0 A0A158NBW0 A0A146LV81 A0A212EHB8 U5EXR1 N6TK58 A0A0L7QTT4 A0A0M9A692 A0A067RT26 A0A1B6FS83 A0A1A9XTA1 A0A1A9V4Y1 A0A1B0CGH0
PDB
3X1D
E-value=0,
Score=1901
Ontologies
GO
GO:0005525
GO:0003924
GO:0016021
GO:0051260
GO:0007029
GO:0031227
GO:0031114
GO:0007517
GO:0042803
GO:0008582
GO:0016320
GO:0005874
GO:0005794
GO:0048691
GO:0051124
GO:0007019
GO:0007030
GO:0061025
GO:0032561
GO:0005737
GO:0012505
GO:0007528
GO:0000139
GO:0005783
GO:0016020
GO:0042802
GO:0015031
GO:0006886
GO:0016192
GO:0030117
GO:0007154
GO:0005112
GO:0015930
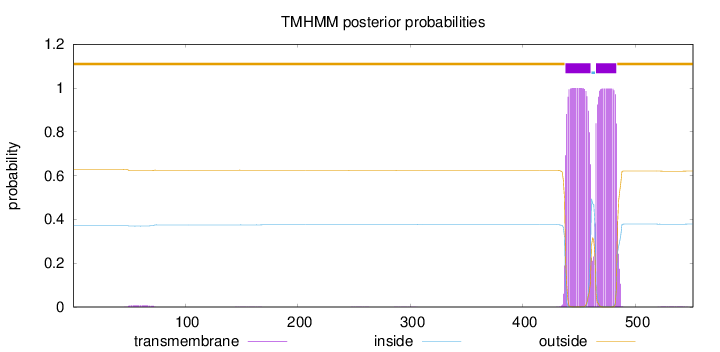
Topology
Subcellular location
Endoplasmic reticulum membrane
Golgi apparatus membrane
Golgi apparatus membrane
Length:
551
Number of predicted TMHs:
2
Exp number of AAs in TMHs:
42.7457700000001
Exp number, first 60 AAs:
0.07813
Total prob of N-in:
0.37136
outside
1 - 437
TMhelix
438 - 460
inside
461 - 464
TMhelix
465 - 483
outside
484 - 551
Population Genetic Test Statistics
Pi
231.492391
Theta
158.941265
Tajima's D
1.673696
CLR
0.109186
CSRT
0.826158692065397
Interpretation
Uncertain
