Pre Gene Modal
BGIBMGA000801
Annotation
PREDICTED:_peptidyl-prolyl_cis-trans_isomerase_FKBP65-like_[Amyelois_transitella]
Full name
Peptidylprolyl isomerase
Location in the cell
Cytoplasmic Reliability : 2.991
Sequence
CDS
ATGCTAGAAGAAGTTATAGTGAGCTCGCGTCCTGATCGGGTTATTCGCAAAAAACTAAAAAATCCAGGTGACTACACAATAGTTCCTTACGAGGATTCTCGTTGCAAGATCAAAATAACAAACTTAGCTTGCACCAACCATCTCGGGAAATGTGAAATTGAACCAGAAAGCAAAGTTTTCAGCACCAATTTCGACGGTAACGTACTAATTGGCGATTCAGATTTTTTTATTGACAAAGACTTTGAGTTAATCCTGCAGCAGATGTGCAGCGGAGAAACTTGTGAAGTGAACATGGTGTATAGGGATGGTAGTGGAGAGCTGGTGAAAGAGATCTCGTGTGACGTAGAGCTGAAGGAAGTGACTGAAGAGCAGCTCATTAGTGACTGGAGCTGGGAGAGATTGATGGAAGCTTCTATACACCACAAGGAGAAAGGTGTAGAATTAGTGAAAGAGAAGCGAATATTAGATGCATTCAGGAGATTCAGTAAGGCTCTCAAGATGGTTGTAGCCATTGAACCTATCGATCCTGAAATCATTGATGAGGAAAAAGCAGTTGCAATTACAGATCTTAAGGTCAAATTATACAATAATTTAGCGCATTGTCAACTTCAATACAATGAATACGAAGCAGCATTAGACCTGTGTAATAGAGCCATTAAATATGACCCAGAAAATGTGAAGGCTCTATATAGAAGATCGCTGGCGTACACCGGCCTGGAGATGTATGTGGAGGCTTGGGCAGACATACAGCGCGTATTACAAATCACACCGGCCGAGAAAGCAGCTGTACAACAATCAAGGCATTTGAAACCGATAATTGATAGGATTAATGAAGATTACAGTAGTGTCGTTAAGAAAATGTTCTCATGA
Protein
MLEEVIVSSRPDRVIRKKLKNPGDYTIVPYEDSRCKIKITNLACTNHLGKCEIEPESKVFSTNFDGNVLIGDSDFFIDKDFELILQQMCSGETCEVNMVYRDGSGELVKEISCDVELKEVTEEQLISDWSWERLMEASIHHKEKGVELVKEKRILDAFRRFSKALKMVVAIEPIDPEIIDEEKAVAITDLKVKLYNNLAHCQLQYNEYEAALDLCNRAIKYDPENVKALYRRSLAYTGLEMYVEAWADIQRVLQITPAEKAAVQQSRHLKPIIDRINEDYSSVVKKMFS
Summary
Catalytic Activity
[protein]-peptidylproline (omega=180) = [protein]-peptidylproline (omega=0)
Uniprot
H9IU71
A0A3S2M2Y9
A0A2W1BP22
A0A2A4IY96
A0A2H1WM93
A0A212EHB1
+ More
A0A194PFQ6 A0A194RHK2 A0A067RVR2 A0A2J7R561 A0A1Y1KX55 A0A2P8YGL9 A0A0N0BHV3 A0A195C9I9 A0A0L7QTU7 A0A232FA50 A0A1B6FPK8 A0A1B6F7G4 K7IUU4 A0A154P472 A0A151X9K9 V5GC97 A0A310S7N3 A0A151J954 A0A158NBV8 A0A0J7KC95 A0A2A3ETS3 A0A195BG58 A0A3L8DT61 E9J8J1 H3AXW6 C3Z4W5 A0A195FDF0 E2B5K7 F4X8E6 E2AR15 A0A1B6LEZ1 J9LTX0 A0A1W0X6S5 V9L945 V9L258 A0A224XHR0 A0A023F6Y9 A0A1B6CQ44 A0A069DR83 A0A2R7X1V6 A0A0V0GCI2 A0A1D1VEV4 A0A0A9X252 A0A146M8T6 R4FL77 V9LAN2 A0A1D2MFY4 V9LDE9 U4U6S2 V9LEY9 V9LEA9 A0A2B4RG75 N6UE90 A0A0P6H634 A0A0P6ING8 A0A369RWD9 R7TMP0 A0A0N7Z8R5 A0A226DCV4 A0A0L8I5K3 A0A1S3JRE7 A0A1S3HJH2 A0A3M6TPX0 B3SDG1 T1IY66 Q641F7 A0A1I8PXB3 A0A1I8PX69
A0A194PFQ6 A0A194RHK2 A0A067RVR2 A0A2J7R561 A0A1Y1KX55 A0A2P8YGL9 A0A0N0BHV3 A0A195C9I9 A0A0L7QTU7 A0A232FA50 A0A1B6FPK8 A0A1B6F7G4 K7IUU4 A0A154P472 A0A151X9K9 V5GC97 A0A310S7N3 A0A151J954 A0A158NBV8 A0A0J7KC95 A0A2A3ETS3 A0A195BG58 A0A3L8DT61 E9J8J1 H3AXW6 C3Z4W5 A0A195FDF0 E2B5K7 F4X8E6 E2AR15 A0A1B6LEZ1 J9LTX0 A0A1W0X6S5 V9L945 V9L258 A0A224XHR0 A0A023F6Y9 A0A1B6CQ44 A0A069DR83 A0A2R7X1V6 A0A0V0GCI2 A0A1D1VEV4 A0A0A9X252 A0A146M8T6 R4FL77 V9LAN2 A0A1D2MFY4 V9LDE9 U4U6S2 V9LEY9 V9LEA9 A0A2B4RG75 N6UE90 A0A0P6H634 A0A0P6ING8 A0A369RWD9 R7TMP0 A0A0N7Z8R5 A0A226DCV4 A0A0L8I5K3 A0A1S3JRE7 A0A1S3HJH2 A0A3M6TPX0 B3SDG1 T1IY66 Q641F7 A0A1I8PXB3 A0A1I8PX69
EC Number
5.2.1.8
Pubmed
EMBL
BABH01034201
RSAL01000057
RVE49772.1
KZ149945
PZC76802.1
NWSH01004562
+ More
PCG64927.1 ODYU01009603 SOQ54148.1 AGBW02014918 OWR40877.1 KQ459604 KPI92122.1 KQ460205 KPJ17047.1 KK852433 KDR23964.1 NEVH01007395 PNF35974.1 GEZM01075111 GEZM01075110 JAV64275.1 PYGN01000611 PSN43383.1 KQ435741 KOX76784.1 KQ978068 KYM97529.1 KQ414740 KOC62078.1 NNAY01000550 OXU27734.1 GECZ01017659 GECZ01002160 JAS52110.1 JAS67609.1 GECZ01029322 GECZ01023976 JAS40447.1 JAS45793.1 AAZX01002640 KQ434809 KZC06647.1 KQ982373 KYQ57053.1 GALX01000696 JAB67770.1 KQ765379 OAD53942.1 KQ979433 KYN21564.1 ADTU01011331 LBMM01009835 KMQ87854.1 KZ288191 PBC34451.1 KQ976500 KYM83150.1 QOIP01000004 RLU23571.1 GL769034 EFZ10866.1 AFYH01118140 GG666582 EEN52322.1 KQ981673 KYN38406.1 GL445852 EFN89019.1 GL888932 EGI57149.1 GL441846 EFN64161.1 GEBQ01030784 GEBQ01017873 JAT09193.1 JAT22104.1 ABLF02019633 MTYJ01000014 OQV23080.1 JW875823 AFP08340.1 JW872506 AFP05024.1 GFTR01004399 JAW12027.1 GBBI01001561 JAC17151.1 GEDC01021662 JAS15636.1 GBGD01002519 JAC86370.1 KK856453 PTY25787.1 GECL01001047 JAP05077.1 BDGG01000005 GAU99440.1 GBHO01028797 GBRD01015047 JAG14807.1 JAG50779.1 GDHC01002461 JAQ16168.1 ACPB03018640 GAHY01002204 JAA75306.1 JW876247 AFP08764.1 LJIJ01001415 ODM91802.1 JW878268 AFP10785.1 KB632095 ERL88767.1 JW878297 AFP10814.1 JW878341 AFP10858.1 LSMT01000603 PFX15843.1 APGK01025879 KB740605 ENN80030.1 GDIQ01024718 JAN70019.1 GDIQ01019053 JAN75684.1 NOWV01000152 RDD38997.1 AMQN01002774 KB310004 ELT92806.1 GDKW01002611 JAI53984.1 LNIX01000025 OXA42768.1 KQ416495 KOF96756.1 RCHS01003197 RMX43457.1 DS985279 EDV19212.1 JH431671 BC082380 AAH82380.1
PCG64927.1 ODYU01009603 SOQ54148.1 AGBW02014918 OWR40877.1 KQ459604 KPI92122.1 KQ460205 KPJ17047.1 KK852433 KDR23964.1 NEVH01007395 PNF35974.1 GEZM01075111 GEZM01075110 JAV64275.1 PYGN01000611 PSN43383.1 KQ435741 KOX76784.1 KQ978068 KYM97529.1 KQ414740 KOC62078.1 NNAY01000550 OXU27734.1 GECZ01017659 GECZ01002160 JAS52110.1 JAS67609.1 GECZ01029322 GECZ01023976 JAS40447.1 JAS45793.1 AAZX01002640 KQ434809 KZC06647.1 KQ982373 KYQ57053.1 GALX01000696 JAB67770.1 KQ765379 OAD53942.1 KQ979433 KYN21564.1 ADTU01011331 LBMM01009835 KMQ87854.1 KZ288191 PBC34451.1 KQ976500 KYM83150.1 QOIP01000004 RLU23571.1 GL769034 EFZ10866.1 AFYH01118140 GG666582 EEN52322.1 KQ981673 KYN38406.1 GL445852 EFN89019.1 GL888932 EGI57149.1 GL441846 EFN64161.1 GEBQ01030784 GEBQ01017873 JAT09193.1 JAT22104.1 ABLF02019633 MTYJ01000014 OQV23080.1 JW875823 AFP08340.1 JW872506 AFP05024.1 GFTR01004399 JAW12027.1 GBBI01001561 JAC17151.1 GEDC01021662 JAS15636.1 GBGD01002519 JAC86370.1 KK856453 PTY25787.1 GECL01001047 JAP05077.1 BDGG01000005 GAU99440.1 GBHO01028797 GBRD01015047 JAG14807.1 JAG50779.1 GDHC01002461 JAQ16168.1 ACPB03018640 GAHY01002204 JAA75306.1 JW876247 AFP08764.1 LJIJ01001415 ODM91802.1 JW878268 AFP10785.1 KB632095 ERL88767.1 JW878297 AFP10814.1 JW878341 AFP10858.1 LSMT01000603 PFX15843.1 APGK01025879 KB740605 ENN80030.1 GDIQ01024718 JAN70019.1 GDIQ01019053 JAN75684.1 NOWV01000152 RDD38997.1 AMQN01002774 KB310004 ELT92806.1 GDKW01002611 JAI53984.1 LNIX01000025 OXA42768.1 KQ416495 KOF96756.1 RCHS01003197 RMX43457.1 DS985279 EDV19212.1 JH431671 BC082380 AAH82380.1
Proteomes
UP000005204
UP000283053
UP000218220
UP000007151
UP000053268
UP000053240
+ More
UP000027135 UP000235965 UP000245037 UP000053105 UP000078542 UP000053825 UP000215335 UP000002358 UP000076502 UP000075809 UP000078492 UP000005205 UP000036403 UP000242457 UP000078540 UP000279307 UP000008672 UP000001554 UP000078541 UP000008237 UP000007755 UP000000311 UP000007819 UP000186922 UP000015103 UP000094527 UP000030742 UP000225706 UP000019118 UP000253843 UP000014760 UP000198287 UP000053454 UP000085678 UP000275408 UP000009022 UP000095300
UP000027135 UP000235965 UP000245037 UP000053105 UP000078542 UP000053825 UP000215335 UP000002358 UP000076502 UP000075809 UP000078492 UP000005205 UP000036403 UP000242457 UP000078540 UP000279307 UP000008672 UP000001554 UP000078541 UP000008237 UP000007755 UP000000311 UP000007819 UP000186922 UP000015103 UP000094527 UP000030742 UP000225706 UP000019118 UP000253843 UP000014760 UP000198287 UP000053454 UP000085678 UP000275408 UP000009022 UP000095300
PRIDE
Pfam
Interpro
IPR040478
FKBP_N_2
+ More
IPR013026 TPR-contain_dom
IPR001440 TPR_1
IPR019734 TPR_repeat
IPR011990 TPR-like_helical_dom_sf
IPR030386 G_GB1_RHD3_dom
IPR015894 Guanylate-bd_N
IPR036543 Guanylate-bd_C_sf
IPR027417 P-loop_NTPase
IPR003191 Guanylate-bd/ATL_C
IPR013105 TPR_2
IPR007853 Znf_DNL-typ
IPR039663 AIP/AIPL1
IPR002347 SDR_fam
IPR036291 NAD(P)-bd_dom_sf
IPR001179 PPIase_FKBP_dom
IPR002048 EF_hand_dom
IPR011992 EF-hand-dom_pair
IPR018864 Nucleoporin_Nup188
IPR018247 EF_Hand_1_Ca_BS
IPR029000 Cyclophilin-like_dom_sf
IPR002130 Cyclophilin-type_PPIase_dom
IPR023566 PPIase_FKBP
IPR031212 FKBP4
IPR013026 TPR-contain_dom
IPR001440 TPR_1
IPR019734 TPR_repeat
IPR011990 TPR-like_helical_dom_sf
IPR030386 G_GB1_RHD3_dom
IPR015894 Guanylate-bd_N
IPR036543 Guanylate-bd_C_sf
IPR027417 P-loop_NTPase
IPR003191 Guanylate-bd/ATL_C
IPR013105 TPR_2
IPR007853 Znf_DNL-typ
IPR039663 AIP/AIPL1
IPR002347 SDR_fam
IPR036291 NAD(P)-bd_dom_sf
IPR001179 PPIase_FKBP_dom
IPR002048 EF_hand_dom
IPR011992 EF-hand-dom_pair
IPR018864 Nucleoporin_Nup188
IPR018247 EF_Hand_1_Ca_BS
IPR029000 Cyclophilin-like_dom_sf
IPR002130 Cyclophilin-type_PPIase_dom
IPR023566 PPIase_FKBP
IPR031212 FKBP4
SUPFAM
Gene 3D
ProteinModelPortal
H9IU71
A0A3S2M2Y9
A0A2W1BP22
A0A2A4IY96
A0A2H1WM93
A0A212EHB1
+ More
A0A194PFQ6 A0A194RHK2 A0A067RVR2 A0A2J7R561 A0A1Y1KX55 A0A2P8YGL9 A0A0N0BHV3 A0A195C9I9 A0A0L7QTU7 A0A232FA50 A0A1B6FPK8 A0A1B6F7G4 K7IUU4 A0A154P472 A0A151X9K9 V5GC97 A0A310S7N3 A0A151J954 A0A158NBV8 A0A0J7KC95 A0A2A3ETS3 A0A195BG58 A0A3L8DT61 E9J8J1 H3AXW6 C3Z4W5 A0A195FDF0 E2B5K7 F4X8E6 E2AR15 A0A1B6LEZ1 J9LTX0 A0A1W0X6S5 V9L945 V9L258 A0A224XHR0 A0A023F6Y9 A0A1B6CQ44 A0A069DR83 A0A2R7X1V6 A0A0V0GCI2 A0A1D1VEV4 A0A0A9X252 A0A146M8T6 R4FL77 V9LAN2 A0A1D2MFY4 V9LDE9 U4U6S2 V9LEY9 V9LEA9 A0A2B4RG75 N6UE90 A0A0P6H634 A0A0P6ING8 A0A369RWD9 R7TMP0 A0A0N7Z8R5 A0A226DCV4 A0A0L8I5K3 A0A1S3JRE7 A0A1S3HJH2 A0A3M6TPX0 B3SDG1 T1IY66 Q641F7 A0A1I8PXB3 A0A1I8PX69
A0A194PFQ6 A0A194RHK2 A0A067RVR2 A0A2J7R561 A0A1Y1KX55 A0A2P8YGL9 A0A0N0BHV3 A0A195C9I9 A0A0L7QTU7 A0A232FA50 A0A1B6FPK8 A0A1B6F7G4 K7IUU4 A0A154P472 A0A151X9K9 V5GC97 A0A310S7N3 A0A151J954 A0A158NBV8 A0A0J7KC95 A0A2A3ETS3 A0A195BG58 A0A3L8DT61 E9J8J1 H3AXW6 C3Z4W5 A0A195FDF0 E2B5K7 F4X8E6 E2AR15 A0A1B6LEZ1 J9LTX0 A0A1W0X6S5 V9L945 V9L258 A0A224XHR0 A0A023F6Y9 A0A1B6CQ44 A0A069DR83 A0A2R7X1V6 A0A0V0GCI2 A0A1D1VEV4 A0A0A9X252 A0A146M8T6 R4FL77 V9LAN2 A0A1D2MFY4 V9LDE9 U4U6S2 V9LEY9 V9LEA9 A0A2B4RG75 N6UE90 A0A0P6H634 A0A0P6ING8 A0A369RWD9 R7TMP0 A0A0N7Z8R5 A0A226DCV4 A0A0L8I5K3 A0A1S3JRE7 A0A1S3HJH2 A0A3M6TPX0 B3SDG1 T1IY66 Q641F7 A0A1I8PXB3 A0A1I8PX69
PDB
1IIP
E-value=2.78057e-12,
Score=172
Ontologies
GO
PANTHER
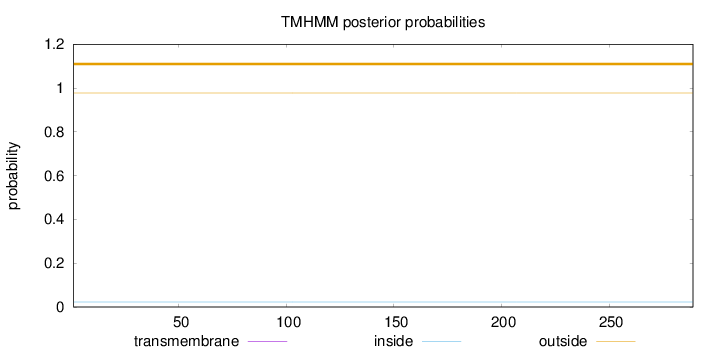
Topology
Length:
289
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00037
Exp number, first 60 AAs:
0
Total prob of N-in:
0.02246
outside
1 - 289
Population Genetic Test Statistics
Pi
189.125801
Theta
153.341744
Tajima's D
0.734906
CLR
0.140987
CSRT
0.584120793960302
Interpretation
Uncertain
