Gene
KWMTBOMO14933
Pre Gene Modal
BGIBMGA000768
Annotation
PREDICTED:_uncharacterized_protein_LOC101745283_isoform_X2_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.035
Sequence
CDS
ATGGCATTATTATTGAAGAGCGTTTGGAGAAACGCACTACGCAACACTCCAATTTGCCAAACAAGACGACACACTAGAAGTCTAACATCGAGCTTAACTGAAGTGAAACATACAAAGACACCAGATCCCACAATGATGAGTTTGAAGAATGCTGTAACTCCTGGTGATGTTATGGGCTTAGTTCAGTCAAATTTACCACAAATGACTGATCAGCATATGCTGCAGGCTTTAAGAAGTCTCTTTCAATTACAGAAGAACAGCAATATTGAAAACTCTGAAGCGGTCATCAAGGATCCAGCATTTGGATTACTCTGTCAGCATTTCAAGAAGCATGCTCGACAGCTTGACGTCAACGAAACAATTGAAGCAATGAAAGTTCTATCATACCTTGGAGTCTCTGTGAATAGTCTAATAGTCCAAACCCTATTACAAATCATAAGATGCGAGATAAACTCTATTGGCATTCGACAGATAATATTCTTAGATTTTATTTTGCATCGATTCAAAGAGAAAAATCATTTAGTTGAAGCTCTCAAAATGGCTTTGCCATTAGCATTTCAAATACATCTACCCTTAGAAATTGATAACGATGATATGCAGTTACTCAAAGATATGTTAGCATACTGTTGCCACCACAATCTTCCAGATCGTTGTATTAATAATGTTGTAGCTGGCCTGTTGCTCCATGACCAAGATCCTGCAAGTGCCAGGTCCATTGTCATGTATTTAAGTTATGTGAATTGTACTGAAGAGGTTTACCCTACCAGGGCTAATTTGCTCAAAGGTTGCTTTAGTGTACTGGCTGAAAATATGGACGATTTACAGTACAATGAAGTTGTGAAGGTGGCTAGTAGAGTAAAGAGTCGTATATTGGATAAGCACCCTGAATTTTATCATGAACAAATGCTGGATGCTATGGGTGATTATGTAATTAAGCACAAAGTGGGTTTTGAGAAGGCATTGCTGATTGCCAGGATATTTTCGAGGATAGCACACACCCACTTAGGACTCGTCCAGTACCTCTGCGAACTGGCAGCATCCAGCCCCACAACTTTATCAGAAGCTCGCACCAACATACTCTTTGGCTTCATCAACTGCCTATCCAACATGAACTACATTCCACCTCCCCCGCAGTGGCAAGAAATCCAAAGACAGATAACCACTAATGCTGTTCTAGATAGTAACAACTCCATATTACCCTGGCCTAAGGTGTGCCTTGAACTGGCTTCGTTAGGGATCTACAAAGACAAAGTCATAAAACATGTGTTCTCAGAGGAGTTTTTAAATGACTATTTCAGTAGAGATCACAATAAGATATTAGATTACATGCAATTGTTTATACTCCGTATAGCAACAAAGACTTTACACGGTGTGGAATATGAGCTCTCTACTGATAGGTTTGGGGGCTCCTGA
Protein
MALLLKSVWRNALRNTPICQTRRHTRSLTSSLTEVKHTKTPDPTMMSLKNAVTPGDVMGLVQSNLPQMTDQHMLQALRSLFQLQKNSNIENSEAVIKDPAFGLLCQHFKKHARQLDVNETIEAMKVLSYLGVSVNSLIVQTLLQIIRCEINSIGIRQIIFLDFILHRFKEKNHLVEALKMALPLAFQIHLPLEIDNDDMQLLKDMLAYCCHHNLPDRCINNVVAGLLLHDQDPASARSIVMYLSYVNCTEEVYPTRANLLKGCFSVLAENMDDLQYNEVVKVASRVKSRILDKHPEFYHEQMLDAMGDYVIKHKVGFEKALLIARIFSRIAHTHLGLVQYLCELAASSPTTLSEARTNILFGFINCLSNMNYIPPPPQWQEIQRQITTNAVLDSNNSILPWPKVCLELASLGIYKDKVIKHVFSEEFLNDYFSRDHNKILDYMQLFILRIATKTLHGVEYELSTDRFGGS
Summary
Uniprot
H9IU38
A0A2W1BR48
A0A2A4IZL1
A0A194RGV2
A0A194PGG6
S4PP42
+ More
A0A2H1WMA6 A0A212EHB7 A0A0L7KXN5 A0A067R3H2 A0A2P8XFL3 A0A2J7QCW9 A0A336MCK0 A0A182VB59 A7USU2 A0A182HSK5 A0A182XAR3 A0A182PCD3 A0A182UFR5 A0A2M4BHM4 A0A2M4BHX8 A0A182F9B6 A0A2J7QCW7 A0A084WT90 W5J8S7 A0A182KE78 A0A023EXU6 A0A182Y6I6 A0A182T1V1 A0A0V0G9N1 A0A224XRK6 A0A182NRH6 A0A1B6DHY0 A0A1B6D2J1 A0A182RLE8 A0A182M1N2 D6WP48 A0A1Y1NDN7 T1IEB1 B0X1I2 A0A1Q3F273 A0A182W0W8 J9JXR6 Q17N49 A0A1B6ET45 A0A1B6HNH2 A0A1B6CUF4 A0A1W4WG44 A0A0A9VVG1 A0A1B6EDX0 N6UCP3 U4UZK5 A0A0K8TK04 A0A1W4WS22 A0A1S3D406 A0A1J1IEV5 A0A0T6B6H5 A0A3L8D9M0 A0A026WXD8 A0A1B6LE44 A0A232FE69 K7J5J5 A0A0P6D6K0 A0A0P6DMU4 A0A0P5RGB2 A0A0N8D826 A0A0P5HWZ5 A0A0P5YD85 A0A0P5SWA3 A0A0P4XZY7 A0A0P5DS38 A0A0N8CN50 E2C817 A0A0P5SYR2 A0A0P5QZ34 A0A0P5IQQ6 A0A0P5E3L5 A0A0N8D3L9 A0A0P5T1D3 A0A0P5L7E8 A0A0N8D432 A0A164K5B5 A0A0P5IW58 A0A0P6I8P1 A0A2S2Q5R2 A0A195CD34 A0A195DUU4 A0A0P6ESU8 A0A151WP12 A0A0P5Z689 A0A0P5BGG9 E9IUM6
A0A2H1WMA6 A0A212EHB7 A0A0L7KXN5 A0A067R3H2 A0A2P8XFL3 A0A2J7QCW9 A0A336MCK0 A0A182VB59 A7USU2 A0A182HSK5 A0A182XAR3 A0A182PCD3 A0A182UFR5 A0A2M4BHM4 A0A2M4BHX8 A0A182F9B6 A0A2J7QCW7 A0A084WT90 W5J8S7 A0A182KE78 A0A023EXU6 A0A182Y6I6 A0A182T1V1 A0A0V0G9N1 A0A224XRK6 A0A182NRH6 A0A1B6DHY0 A0A1B6D2J1 A0A182RLE8 A0A182M1N2 D6WP48 A0A1Y1NDN7 T1IEB1 B0X1I2 A0A1Q3F273 A0A182W0W8 J9JXR6 Q17N49 A0A1B6ET45 A0A1B6HNH2 A0A1B6CUF4 A0A1W4WG44 A0A0A9VVG1 A0A1B6EDX0 N6UCP3 U4UZK5 A0A0K8TK04 A0A1W4WS22 A0A1S3D406 A0A1J1IEV5 A0A0T6B6H5 A0A3L8D9M0 A0A026WXD8 A0A1B6LE44 A0A232FE69 K7J5J5 A0A0P6D6K0 A0A0P6DMU4 A0A0P5RGB2 A0A0N8D826 A0A0P5HWZ5 A0A0P5YD85 A0A0P5SWA3 A0A0P4XZY7 A0A0P5DS38 A0A0N8CN50 E2C817 A0A0P5SYR2 A0A0P5QZ34 A0A0P5IQQ6 A0A0P5E3L5 A0A0N8D3L9 A0A0P5T1D3 A0A0P5L7E8 A0A0N8D432 A0A164K5B5 A0A0P5IW58 A0A0P6I8P1 A0A2S2Q5R2 A0A195CD34 A0A195DUU4 A0A0P6ESU8 A0A151WP12 A0A0P5Z689 A0A0P5BGG9 E9IUM6
Pubmed
EMBL
BABH01034200
BABH01034201
KZ149945
PZC76801.1
NWSH01004562
PCG64926.1
+ More
KQ460205 KPJ17048.1 KQ459604 KPI92123.1 GAIX01000452 JAA92108.1 ODYU01009603 SOQ54147.1 AGBW02014918 OWR40876.1 JTDY01004664 KOB67900.1 KK852934 KDR13615.1 PYGN01002315 PSN30798.1 NEVH01015858 PNF26430.1 UFQT01000380 SSX23748.1 AAAB01008859 EDO64227.2 APCN01000282 GGFJ01003419 MBW52560.1 GGFJ01003420 MBW52561.1 PNF26429.1 ATLV01026872 KE525420 KFB53434.1 ADMH02002099 ETN59204.1 GBBI01004607 JAC14105.1 GECL01001398 JAP04726.1 GFTR01005817 JAW10609.1 GEDC01017899 GEDC01012020 JAS19399.1 JAS25278.1 GEDC01017369 JAS19929.1 AXCM01002550 KQ971352 EFA07285.1 GEZM01009725 GEZM01009724 JAV94356.1 ACPB03017593 DS232260 EDS38631.1 GFDL01013404 JAV21641.1 ABLF02037217 CH477202 EAT48094.1 EAT48095.1 GECZ01028876 JAS40893.1 GECU01031469 JAS76237.1 GEDC01023072 GEDC01020129 GEDC01010463 JAS14226.1 JAS17169.1 JAS26835.1 GBHO01044423 GDHC01004803 JAF99180.1 JAQ13826.1 GEDC01001173 JAS36125.1 APGK01028591 KB740694 ENN79435.1 KI208145 KI210265 ERL95805.1 ERL96216.1 GBRD01000211 JAG65610.1 CVRI01000048 CRK98747.1 LJIG01009502 KRT82952.1 QOIP01000011 RLU16842.1 KK107078 EZA60498.1 GEBQ01018213 JAT21764.1 NNAY01000389 OXU28729.1 GDIQ01089941 JAN04796.1 GDIQ01089942 JAN04795.1 GDIQ01110490 JAL41236.1 GDIP01059639 JAM44076.1 GDIQ01221796 JAK29929.1 GDIP01061484 JAM42231.1 GDIP01141651 JAL62063.1 GDIP01235632 JAI87769.1 GDIP01153624 JAJ69778.1 GDIP01115447 JAL88267.1 GL453506 EFN75935.1 GDIP01133489 JAL70225.1 GDIQ01113593 JAL38133.1 GDIQ01211698 JAK40027.1 GDIP01147538 JAJ75864.1 GDIP01072095 JAM31620.1 GDIP01134834 JAL68880.1 GDIQ01175562 JAK76163.1 GDIP01070791 JAM32924.1 LRGB01003375 KZS02959.1 GDIQ01209694 JAK42031.1 GDIQ01007969 JAN86768.1 GGMS01003874 MBY73077.1 KQ978023 KYM97993.1 KQ980322 KYN16616.1 GDIQ01071847 JAN22890.1 KQ982888 KYQ49642.1 GDIP01049140 JAM54575.1 GDIP01184978 JAJ38424.1 GL765999 EFZ15720.1
KQ460205 KPJ17048.1 KQ459604 KPI92123.1 GAIX01000452 JAA92108.1 ODYU01009603 SOQ54147.1 AGBW02014918 OWR40876.1 JTDY01004664 KOB67900.1 KK852934 KDR13615.1 PYGN01002315 PSN30798.1 NEVH01015858 PNF26430.1 UFQT01000380 SSX23748.1 AAAB01008859 EDO64227.2 APCN01000282 GGFJ01003419 MBW52560.1 GGFJ01003420 MBW52561.1 PNF26429.1 ATLV01026872 KE525420 KFB53434.1 ADMH02002099 ETN59204.1 GBBI01004607 JAC14105.1 GECL01001398 JAP04726.1 GFTR01005817 JAW10609.1 GEDC01017899 GEDC01012020 JAS19399.1 JAS25278.1 GEDC01017369 JAS19929.1 AXCM01002550 KQ971352 EFA07285.1 GEZM01009725 GEZM01009724 JAV94356.1 ACPB03017593 DS232260 EDS38631.1 GFDL01013404 JAV21641.1 ABLF02037217 CH477202 EAT48094.1 EAT48095.1 GECZ01028876 JAS40893.1 GECU01031469 JAS76237.1 GEDC01023072 GEDC01020129 GEDC01010463 JAS14226.1 JAS17169.1 JAS26835.1 GBHO01044423 GDHC01004803 JAF99180.1 JAQ13826.1 GEDC01001173 JAS36125.1 APGK01028591 KB740694 ENN79435.1 KI208145 KI210265 ERL95805.1 ERL96216.1 GBRD01000211 JAG65610.1 CVRI01000048 CRK98747.1 LJIG01009502 KRT82952.1 QOIP01000011 RLU16842.1 KK107078 EZA60498.1 GEBQ01018213 JAT21764.1 NNAY01000389 OXU28729.1 GDIQ01089941 JAN04796.1 GDIQ01089942 JAN04795.1 GDIQ01110490 JAL41236.1 GDIP01059639 JAM44076.1 GDIQ01221796 JAK29929.1 GDIP01061484 JAM42231.1 GDIP01141651 JAL62063.1 GDIP01235632 JAI87769.1 GDIP01153624 JAJ69778.1 GDIP01115447 JAL88267.1 GL453506 EFN75935.1 GDIP01133489 JAL70225.1 GDIQ01113593 JAL38133.1 GDIQ01211698 JAK40027.1 GDIP01147538 JAJ75864.1 GDIP01072095 JAM31620.1 GDIP01134834 JAL68880.1 GDIQ01175562 JAK76163.1 GDIP01070791 JAM32924.1 LRGB01003375 KZS02959.1 GDIQ01209694 JAK42031.1 GDIQ01007969 JAN86768.1 GGMS01003874 MBY73077.1 KQ978023 KYM97993.1 KQ980322 KYN16616.1 GDIQ01071847 JAN22890.1 KQ982888 KYQ49642.1 GDIP01049140 JAM54575.1 GDIP01184978 JAJ38424.1 GL765999 EFZ15720.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000007151
UP000037510
+ More
UP000027135 UP000245037 UP000235965 UP000075903 UP000007062 UP000075840 UP000076407 UP000075885 UP000075902 UP000069272 UP000030765 UP000000673 UP000075881 UP000076408 UP000075901 UP000075884 UP000075900 UP000075883 UP000007266 UP000015103 UP000002320 UP000075920 UP000007819 UP000008820 UP000192223 UP000019118 UP000030742 UP000079169 UP000183832 UP000279307 UP000053097 UP000215335 UP000002358 UP000008237 UP000076858 UP000078542 UP000078492 UP000075809
UP000027135 UP000245037 UP000235965 UP000075903 UP000007062 UP000075840 UP000076407 UP000075885 UP000075902 UP000069272 UP000030765 UP000000673 UP000075881 UP000076408 UP000075901 UP000075884 UP000075900 UP000075883 UP000007266 UP000015103 UP000002320 UP000075920 UP000007819 UP000008820 UP000192223 UP000019118 UP000030742 UP000079169 UP000183832 UP000279307 UP000053097 UP000215335 UP000002358 UP000008237 UP000076858 UP000078542 UP000078492 UP000075809
PRIDE
Interpro
SUPFAM
SSF48371
SSF48371
ProteinModelPortal
H9IU38
A0A2W1BR48
A0A2A4IZL1
A0A194RGV2
A0A194PGG6
S4PP42
+ More
A0A2H1WMA6 A0A212EHB7 A0A0L7KXN5 A0A067R3H2 A0A2P8XFL3 A0A2J7QCW9 A0A336MCK0 A0A182VB59 A7USU2 A0A182HSK5 A0A182XAR3 A0A182PCD3 A0A182UFR5 A0A2M4BHM4 A0A2M4BHX8 A0A182F9B6 A0A2J7QCW7 A0A084WT90 W5J8S7 A0A182KE78 A0A023EXU6 A0A182Y6I6 A0A182T1V1 A0A0V0G9N1 A0A224XRK6 A0A182NRH6 A0A1B6DHY0 A0A1B6D2J1 A0A182RLE8 A0A182M1N2 D6WP48 A0A1Y1NDN7 T1IEB1 B0X1I2 A0A1Q3F273 A0A182W0W8 J9JXR6 Q17N49 A0A1B6ET45 A0A1B6HNH2 A0A1B6CUF4 A0A1W4WG44 A0A0A9VVG1 A0A1B6EDX0 N6UCP3 U4UZK5 A0A0K8TK04 A0A1W4WS22 A0A1S3D406 A0A1J1IEV5 A0A0T6B6H5 A0A3L8D9M0 A0A026WXD8 A0A1B6LE44 A0A232FE69 K7J5J5 A0A0P6D6K0 A0A0P6DMU4 A0A0P5RGB2 A0A0N8D826 A0A0P5HWZ5 A0A0P5YD85 A0A0P5SWA3 A0A0P4XZY7 A0A0P5DS38 A0A0N8CN50 E2C817 A0A0P5SYR2 A0A0P5QZ34 A0A0P5IQQ6 A0A0P5E3L5 A0A0N8D3L9 A0A0P5T1D3 A0A0P5L7E8 A0A0N8D432 A0A164K5B5 A0A0P5IW58 A0A0P6I8P1 A0A2S2Q5R2 A0A195CD34 A0A195DUU4 A0A0P6ESU8 A0A151WP12 A0A0P5Z689 A0A0P5BGG9 E9IUM6
A0A2H1WMA6 A0A212EHB7 A0A0L7KXN5 A0A067R3H2 A0A2P8XFL3 A0A2J7QCW9 A0A336MCK0 A0A182VB59 A7USU2 A0A182HSK5 A0A182XAR3 A0A182PCD3 A0A182UFR5 A0A2M4BHM4 A0A2M4BHX8 A0A182F9B6 A0A2J7QCW7 A0A084WT90 W5J8S7 A0A182KE78 A0A023EXU6 A0A182Y6I6 A0A182T1V1 A0A0V0G9N1 A0A224XRK6 A0A182NRH6 A0A1B6DHY0 A0A1B6D2J1 A0A182RLE8 A0A182M1N2 D6WP48 A0A1Y1NDN7 T1IEB1 B0X1I2 A0A1Q3F273 A0A182W0W8 J9JXR6 Q17N49 A0A1B6ET45 A0A1B6HNH2 A0A1B6CUF4 A0A1W4WG44 A0A0A9VVG1 A0A1B6EDX0 N6UCP3 U4UZK5 A0A0K8TK04 A0A1W4WS22 A0A1S3D406 A0A1J1IEV5 A0A0T6B6H5 A0A3L8D9M0 A0A026WXD8 A0A1B6LE44 A0A232FE69 K7J5J5 A0A0P6D6K0 A0A0P6DMU4 A0A0P5RGB2 A0A0N8D826 A0A0P5HWZ5 A0A0P5YD85 A0A0P5SWA3 A0A0P4XZY7 A0A0P5DS38 A0A0N8CN50 E2C817 A0A0P5SYR2 A0A0P5QZ34 A0A0P5IQQ6 A0A0P5E3L5 A0A0N8D3L9 A0A0P5T1D3 A0A0P5L7E8 A0A0N8D432 A0A164K5B5 A0A0P5IW58 A0A0P6I8P1 A0A2S2Q5R2 A0A195CD34 A0A195DUU4 A0A0P6ESU8 A0A151WP12 A0A0P5Z689 A0A0P5BGG9 E9IUM6
Ontologies
KEGG
PANTHER
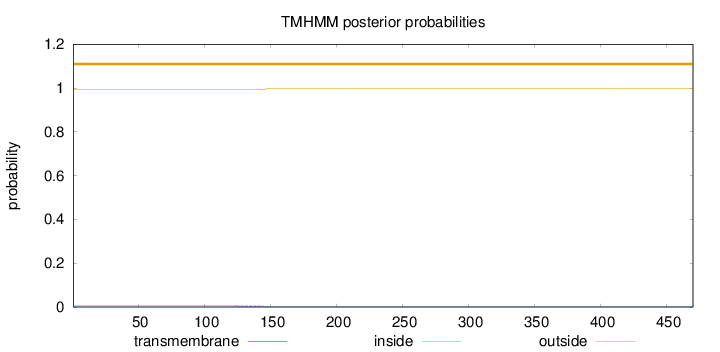
Topology
Length:
470
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.14015
Exp number, first 60 AAs:
0.00015
Total prob of N-in:
0.00822
outside
1 - 470
Population Genetic Test Statistics
Pi
45.75146
Theta
157.068526
Tajima's D
-1.727343
CLR
502.752079
CSRT
0.0348982550872456
Interpretation
Uncertain
