Gene
KWMTBOMO14932
Pre Gene Modal
BGIBMGA000768
Annotation
putative_phospholipase_B-like_2_precursor_[Danaus_plexippus]
Full name
Phospholipase B-like
+ More
Putative phospholipase B-like 2
Putative phospholipase B-like 2
Alternative Name
76 kDa protein
LAMA-like protein 2
Lamina ancestor homolog 2
Phospholipase B domain-containing protein 2
LAMA-like protein 2
Lamina ancestor homolog 2
Phospholipase B domain-containing protein 2
Location in the cell
Lysosomal Reliability : 1.863 PlasmaMembrane Reliability : 1.226
Sequence
CDS
ATGTTGATTAGTTTAATATTTTTTAGTTTATTTTATGCTTGTAAATGTGAATATGGGTTTGTTTTGTGGTCTGAAAATAAACCTACTGTAACCGTGACTGACTATTACGCTGATATACCCGATGTATACGTAGCGAAAGCAACTTACACCAACGAAATAAATAATACAGGTTGGGCATTTCTTGAGCTGCACACATCAAGAGACTCAACAGACGAGAAACAAGCTTATTCAGCTGGTTTTCTGGAAGGATTCCTCACAAGAGATCTGATATGGATGCATTGGCAAAATGTTCTGAAAGGATATTGCTTTAACAAAACTGATGTATGCGCACTAATTGAAGATTATTTAGATAAAAATGAAGATTACATAGCATCTATGGTGCAAGAACACTACGAGGATCCTTATTGGTACCAGATAAAGCTATACTACATCCAGCTGGAAGGCCTAGCAGCCGGCTACAATGATGCCACTAAGAGCATGTACGAGTGGCTCACTGTAAGAGATATACTTTGGATTAACATGCTGGGTGACTTGGACGACCTGGCGTTCGCGCTGTCCATGCCAAGGGAGACGCCCGAGGGCCTGATGTTCGGGGAGCACTGCTCGGGGCTGGTGAAGCTGCTGCCGGACCTCTCGGACCTGTACACGGCGCAGGTCACGTGGAACAGCTACCAGTCGATGCTACGGATCCAAAAGATGTACGTGATGAACTACATGACGTCACCGGGCAGCGGGGTCCTCATACCCGGCAGGAAAATGTCGCTCTCCTCCTACCCGGCCTTCGTACAGTCGACGGACGACTTCTACGTGATGTCGTCGGGGCTGGTGGCCGCCGAGACCACCATCGGCAACAGCAACCGGACGCTGTACGAGCACGTGCGGCCAGAGGGACAGATATTGGAGTTCGTTCGCGCCATGGTCGCCAACCGGCTGGCCACGGACGGCGCGCGCTGGGTCCGCCTGTTCAGGGACCACAACAGCGGCACCTATAACAACCAGTGGTACGTTGTAGATTACAACAAGTTCACGCCGGCTGTTGGGAAGATGAAGGGGTCTGTGAGGCCGGGCCTGCTGTGGGTGGTTGAGCAGCTACCGGGATACACAGAGGCGGCCGATATAACTGAAGAGCTCAAGCAAACTTCGTATTTCCCATCCTACAACATCGCGTACTTCCCGAGCGTCTTCAACTTGAGCGGAGGGAACGAGCGAGTGAACACTTACGGCGACTGGTTCGGCTACCACACCAACCCCAGGGCGAAGATATTCAAGCAGAAACAGAGTGAGATCCACAATCTGCGCGACATGTACCGGACGATGCGGTACAACGACTACCGGCACGACCCGCTGTCGCGCTGCGCCCCCTGCGCGCCCCCCTACAGCGCCGCCAACGCAGTCGCCGCCAGGAACGACCTCAACCCCGCCAACGGTACGTACCCGTTCAGAGCCTTAGGACATCGCAGTCACGGAGCCACCGACGCGAAGATCACGAGCTCGCTGCTCAGGAAGACTTACCAGTTCATAGCCGTGTCGAGCCCCACCTACAACTTGTCCAGGGGGGTGCCGCCCTTCGTTTGGAGTAAGTTCGACATGGGTCCGCAGATATCTCACGTGGGCCACCCCGATGTGTGGATGTTCCCACCCTTGCTCCACCGCTGGGAGTGGGGATAG
Protein
MLISLIFFSLFYACKCEYGFVLWSENKPTVTVTDYYADIPDVYVAKATYTNEINNTGWAFLELHTSRDSTDEKQAYSAGFLEGFLTRDLIWMHWQNVLKGYCFNKTDVCALIEDYLDKNEDYIASMVQEHYEDPYWYQIKLYYIQLEGLAAGYNDATKSMYEWLTVRDILWINMLGDLDDLAFALSMPRETPEGLMFGEHCSGLVKLLPDLSDLYTAQVTWNSYQSMLRIQKMYVMNYMTSPGSGVLIPGRKMSLSSYPAFVQSTDDFYVMSSGLVAAETTIGNSNRTLYEHVRPEGQILEFVRAMVANRLATDGARWVRLFRDHNSGTYNNQWYVVDYNKFTPAVGKMKGSVRPGLLWVVEQLPGYTEAADITEELKQTSYFPSYNIAYFPSVFNLSGGNERVNTYGDWFGYHTNPRAKIFKQKQSEIHNLRDMYRTMRYNDYRHDPLSRCAPCAPPYSAANAVAARNDLNPANGTYPFRALGHRSHGATDAKITSSLLRKTYQFIAVSSPTYNLSRGVPPFVWSKFDMGPQISHVGHPDVWMFPPLLHRWEWG
Summary
Description
Putative phospholipase.
Subunit
Interacts with IGF2R.
Similarity
Belongs to the phospholipase B-like family.
Keywords
Alternative splicing
Complete proteome
Direct protein sequencing
Disulfide bond
Glycoprotein
Hydrolase
Lipid degradation
Lipid metabolism
Lysosome
Polymorphism
Reference proteome
Signal
Feature
chain Phospholipase B-like
splice variant In isoform 2.
sequence variant In dbSNP:rs7965471.
splice variant In isoform 2.
sequence variant In dbSNP:rs7965471.
Uniprot
A0A212EHB7
A0A3S2NYN4
A0A2H1WMA6
A0A1E1VY59
A0A2W1BVI5
A0A194PHD8
+ More
A0A2J7PU65 J9K224 A0A2I9LPG8 A0A2H8TJI2 A0A0P5J9R6 A0A164ZX29 A0A0N8BAQ7 A0A0P6EVS2 A0A0N8B5Q0 A0A0P6GG78 A0A0P5AKM6 A0A0P5UML3 A0A0P5GTM8 A0A0N8DYW5 A0A0P5RGR3 A0A067RJD1 A0A0P5BN34 A0A0P6DC37 A0A0P5HHQ7 A0A0P6AAA5 A0A0P5QS33 A0A0P5FWY2 A0A0P6IZ23 A0A0P5IGE3 A0A0P5HKV5 A0A0P5K8J4 A0A0P5MCI9 E9GSQ2 A0A0N8E886 A0A0P5MSK5 A0A0K2SZE0 A0A0P5QAW2 A0A0P5QP37 A0A0P6FKH2 A0A1W7R9X8 R7VB97 A0A3R7PL47 A0A0P5NEB0 A0A1S3H1M5 A0A0P5JS83 A0A2P8YQZ5 T1IJU9 A0A1S3H3H8 A0A0P6ECV0 A0A0P6IGS1 A0A0P5Q498 A0A2R8QFI9 A0A0P5ZP95 A0A0P5G2M1 A0A0P5H812 A2RV19 A0A293M231 A0A3B3HSJ2 A0A0P6FAS0 V9KJN6 A0A3P9KUP8 A0A0S7G278 A0A3P9H0U9 A0A210QBW5 V9KKP6 H2LVN2 A0A3B1IZJ5 W5U8Z2 M3ZZL5 A0A0P4VWC0 V9KIX4 A0A3B4EI41 J9JWY7 V5H983 A0A034WWT4 A0A1V9XR98 A0A151N4M2 A0A060WGU2 G7PIR3 T1FMY2 G7N5Q2 A0A224YPI2 A0A2K5E2G1 A0A315V885 A0A2K6S5I6 A0A0P5PVF2 A0A2I2UDS4 F7I3N3 A0A0P5DKU7 A0A2K5YWZ4 Q8NHP8 A0A2K6DCS4 A0A2K5WZV8 I2CYI6 A0A131YKS0 U3ECB3 H9YUP8 H9F441
A0A2J7PU65 J9K224 A0A2I9LPG8 A0A2H8TJI2 A0A0P5J9R6 A0A164ZX29 A0A0N8BAQ7 A0A0P6EVS2 A0A0N8B5Q0 A0A0P6GG78 A0A0P5AKM6 A0A0P5UML3 A0A0P5GTM8 A0A0N8DYW5 A0A0P5RGR3 A0A067RJD1 A0A0P5BN34 A0A0P6DC37 A0A0P5HHQ7 A0A0P6AAA5 A0A0P5QS33 A0A0P5FWY2 A0A0P6IZ23 A0A0P5IGE3 A0A0P5HKV5 A0A0P5K8J4 A0A0P5MCI9 E9GSQ2 A0A0N8E886 A0A0P5MSK5 A0A0K2SZE0 A0A0P5QAW2 A0A0P5QP37 A0A0P6FKH2 A0A1W7R9X8 R7VB97 A0A3R7PL47 A0A0P5NEB0 A0A1S3H1M5 A0A0P5JS83 A0A2P8YQZ5 T1IJU9 A0A1S3H3H8 A0A0P6ECV0 A0A0P6IGS1 A0A0P5Q498 A0A2R8QFI9 A0A0P5ZP95 A0A0P5G2M1 A0A0P5H812 A2RV19 A0A293M231 A0A3B3HSJ2 A0A0P6FAS0 V9KJN6 A0A3P9KUP8 A0A0S7G278 A0A3P9H0U9 A0A210QBW5 V9KKP6 H2LVN2 A0A3B1IZJ5 W5U8Z2 M3ZZL5 A0A0P4VWC0 V9KIX4 A0A3B4EI41 J9JWY7 V5H983 A0A034WWT4 A0A1V9XR98 A0A151N4M2 A0A060WGU2 G7PIR3 T1FMY2 G7N5Q2 A0A224YPI2 A0A2K5E2G1 A0A315V885 A0A2K6S5I6 A0A0P5PVF2 A0A2I2UDS4 F7I3N3 A0A0P5DKU7 A0A2K5YWZ4 Q8NHP8 A0A2K6DCS4 A0A2K5WZV8 I2CYI6 A0A131YKS0 U3ECB3 H9YUP8 H9F441
EC Number
3.1.1.-
Pubmed
22118469
28756777
26354079
29248469
24845553
21292972
+ More
23254933 29403074 23594743 17554307 24402279 28812685 25329095 23127152 23542700 25765539 24762651 28327890 22293439 24755649 22002653 28797301 29703783 17975172 16541075 15489334 17105447 19159218 21269460 25944712 25319552 26830274 25243066
23254933 29403074 23594743 17554307 24402279 28812685 25329095 23127152 23542700 25765539 24762651 28327890 22293439 24755649 22002653 28797301 29703783 17975172 16541075 15489334 17105447 19159218 21269460 25944712 25319552 26830274 25243066
EMBL
AGBW02014918
OWR40876.1
RSAL01000022
RVE52351.1
ODYU01009603
SOQ54147.1
+ More
GDQN01011388 JAT79666.1 KZ149945 PZC76800.1 KQ459604 KPI92124.1 NEVH01021208 PNF19877.1 ABLF02016253 ABLF02016254 GFWZ01000288 MBW20278.1 GFXV01002394 MBW14199.1 GDIQ01209012 GDIQ01209011 JAK42713.1 LRGB01000687 KZS16891.1 GDIQ01269525 GDIQ01219257 GDIQ01196203 JAK55522.1 GDIQ01064566 JAN30171.1 GDIQ01210259 JAK41466.1 GDIQ01044063 JAN50674.1 GDIP01197995 JAJ25407.1 GDIP01110890 JAL92824.1 GDIQ01235749 GDIQ01052179 JAK15976.1 GDIQ01078348 JAN16389.1 GDIQ01110335 JAL41391.1 KK852473 KDR23108.1 GDIP01183079 JAJ40323.1 GDIQ01080247 JAN14490.1 GDIQ01233292 GDIQ01125487 JAK18433.1 GDIP01032596 JAM71119.1 GDIQ01110336 JAL41390.1 GDIQ01254598 JAJ97126.1 GDIQ01025520 JAN69217.1 GDIQ01213750 JAK37975.1 GDIQ01231640 JAK20085.1 GDIQ01213749 JAK37976.1 GDIQ01157624 JAK94101.1 GL732562 EFX77541.1 GDIQ01052180 JAN42557.1 GDIQ01152521 JAK99204.1 HACA01001793 CDW19154.1 GDIQ01138384 JAL13342.1 GDIQ01112086 JAL39640.1 GDIQ01063061 JAN31676.1 GFAH01000439 JAV47950.1 AMQN01004460 KB293569 ELU15812.1 QCYY01001783 ROT75354.1 GDIQ01146629 JAL05097.1 GDIQ01200518 JAK51207.1 PYGN01000420 PSN46677.1 JH430359 GDIQ01064567 JAN30170.1 GDIQ01031225 JAN63512.1 GDIQ01140533 JAL11193.1 CABZ01067558 GDIP01040606 JAM63109.1 GDIQ01247434 JAK04291.1 GDIQ01236806 JAK14919.1 BC133139 AAI33140.1 GFWV01010304 MAA35033.1 GDIQ01063062 JAN31675.1 JW865809 AFO98326.1 GBYX01477768 GBYX01458196 GBYX01458194 JAO23359.1 NEDP02004233 OWF46224.1 JW865989 AFO98506.1 JT407599 AHH38044.1 GDRN01103450 JAI58085.1 JW865413 AFO97930.1 ABLF02016250 GANP01013261 JAB71207.1 GBBO01000014 JAC59114.1 MNPL01005424 OQR76026.1 AKHW03004053 KYO31774.1 FR904540 CDQ66372.1 CM001286 EHH66740.1 AMQM01004014 KB096411 ESO04980.1 CM001263 EHH21209.1 GFPF01004594 MAA15740.1 NHOQ01002094 PWA19550.1 GDIQ01129778 JAL21948.1 AANG04002472 GDIP01154796 JAJ68606.1 AC010178 BC030618 BC071832 AQIA01012788 JV667358 AFJ72021.1 GEDV01009455 JAP79102.1 GAMT01002761 GAMS01005770 GAMR01002847 GAMQ01003538 GAMP01010553 JAB09100.1 JAB17366.1 JAB31085.1 JAB38313.1 JAB42202.1 JU470610 AFH27414.1 JU325644 AFE69400.1
GDQN01011388 JAT79666.1 KZ149945 PZC76800.1 KQ459604 KPI92124.1 NEVH01021208 PNF19877.1 ABLF02016253 ABLF02016254 GFWZ01000288 MBW20278.1 GFXV01002394 MBW14199.1 GDIQ01209012 GDIQ01209011 JAK42713.1 LRGB01000687 KZS16891.1 GDIQ01269525 GDIQ01219257 GDIQ01196203 JAK55522.1 GDIQ01064566 JAN30171.1 GDIQ01210259 JAK41466.1 GDIQ01044063 JAN50674.1 GDIP01197995 JAJ25407.1 GDIP01110890 JAL92824.1 GDIQ01235749 GDIQ01052179 JAK15976.1 GDIQ01078348 JAN16389.1 GDIQ01110335 JAL41391.1 KK852473 KDR23108.1 GDIP01183079 JAJ40323.1 GDIQ01080247 JAN14490.1 GDIQ01233292 GDIQ01125487 JAK18433.1 GDIP01032596 JAM71119.1 GDIQ01110336 JAL41390.1 GDIQ01254598 JAJ97126.1 GDIQ01025520 JAN69217.1 GDIQ01213750 JAK37975.1 GDIQ01231640 JAK20085.1 GDIQ01213749 JAK37976.1 GDIQ01157624 JAK94101.1 GL732562 EFX77541.1 GDIQ01052180 JAN42557.1 GDIQ01152521 JAK99204.1 HACA01001793 CDW19154.1 GDIQ01138384 JAL13342.1 GDIQ01112086 JAL39640.1 GDIQ01063061 JAN31676.1 GFAH01000439 JAV47950.1 AMQN01004460 KB293569 ELU15812.1 QCYY01001783 ROT75354.1 GDIQ01146629 JAL05097.1 GDIQ01200518 JAK51207.1 PYGN01000420 PSN46677.1 JH430359 GDIQ01064567 JAN30170.1 GDIQ01031225 JAN63512.1 GDIQ01140533 JAL11193.1 CABZ01067558 GDIP01040606 JAM63109.1 GDIQ01247434 JAK04291.1 GDIQ01236806 JAK14919.1 BC133139 AAI33140.1 GFWV01010304 MAA35033.1 GDIQ01063062 JAN31675.1 JW865809 AFO98326.1 GBYX01477768 GBYX01458196 GBYX01458194 JAO23359.1 NEDP02004233 OWF46224.1 JW865989 AFO98506.1 JT407599 AHH38044.1 GDRN01103450 JAI58085.1 JW865413 AFO97930.1 ABLF02016250 GANP01013261 JAB71207.1 GBBO01000014 JAC59114.1 MNPL01005424 OQR76026.1 AKHW03004053 KYO31774.1 FR904540 CDQ66372.1 CM001286 EHH66740.1 AMQM01004014 KB096411 ESO04980.1 CM001263 EHH21209.1 GFPF01004594 MAA15740.1 NHOQ01002094 PWA19550.1 GDIQ01129778 JAL21948.1 AANG04002472 GDIP01154796 JAJ68606.1 AC010178 BC030618 BC071832 AQIA01012788 JV667358 AFJ72021.1 GEDV01009455 JAP79102.1 GAMT01002761 GAMS01005770 GAMR01002847 GAMQ01003538 GAMP01010553 JAB09100.1 JAB17366.1 JAB31085.1 JAB38313.1 JAB42202.1 JU470610 AFH27414.1 JU325644 AFE69400.1
Proteomes
UP000007151
UP000283053
UP000053268
UP000235965
UP000007819
UP000076858
+ More
UP000027135 UP000000305 UP000014760 UP000283509 UP000085678 UP000245037 UP000000437 UP000001038 UP000265180 UP000265200 UP000242188 UP000018467 UP000221080 UP000002852 UP000261440 UP000192247 UP000050525 UP000193380 UP000009130 UP000015101 UP000233020 UP000233220 UP000011712 UP000008225 UP000233140 UP000005640 UP000233120 UP000233100
UP000027135 UP000000305 UP000014760 UP000283509 UP000085678 UP000245037 UP000000437 UP000001038 UP000265180 UP000265200 UP000242188 UP000018467 UP000221080 UP000002852 UP000261440 UP000192247 UP000050525 UP000193380 UP000009130 UP000015101 UP000233020 UP000233220 UP000011712 UP000008225 UP000233140 UP000005640 UP000233120 UP000233100
PRIDE
Pfam
PF04916 Phospholip_B
Interpro
IPR007000
PLipase_B-like
ProteinModelPortal
A0A212EHB7
A0A3S2NYN4
A0A2H1WMA6
A0A1E1VY59
A0A2W1BVI5
A0A194PHD8
+ More
A0A2J7PU65 J9K224 A0A2I9LPG8 A0A2H8TJI2 A0A0P5J9R6 A0A164ZX29 A0A0N8BAQ7 A0A0P6EVS2 A0A0N8B5Q0 A0A0P6GG78 A0A0P5AKM6 A0A0P5UML3 A0A0P5GTM8 A0A0N8DYW5 A0A0P5RGR3 A0A067RJD1 A0A0P5BN34 A0A0P6DC37 A0A0P5HHQ7 A0A0P6AAA5 A0A0P5QS33 A0A0P5FWY2 A0A0P6IZ23 A0A0P5IGE3 A0A0P5HKV5 A0A0P5K8J4 A0A0P5MCI9 E9GSQ2 A0A0N8E886 A0A0P5MSK5 A0A0K2SZE0 A0A0P5QAW2 A0A0P5QP37 A0A0P6FKH2 A0A1W7R9X8 R7VB97 A0A3R7PL47 A0A0P5NEB0 A0A1S3H1M5 A0A0P5JS83 A0A2P8YQZ5 T1IJU9 A0A1S3H3H8 A0A0P6ECV0 A0A0P6IGS1 A0A0P5Q498 A0A2R8QFI9 A0A0P5ZP95 A0A0P5G2M1 A0A0P5H812 A2RV19 A0A293M231 A0A3B3HSJ2 A0A0P6FAS0 V9KJN6 A0A3P9KUP8 A0A0S7G278 A0A3P9H0U9 A0A210QBW5 V9KKP6 H2LVN2 A0A3B1IZJ5 W5U8Z2 M3ZZL5 A0A0P4VWC0 V9KIX4 A0A3B4EI41 J9JWY7 V5H983 A0A034WWT4 A0A1V9XR98 A0A151N4M2 A0A060WGU2 G7PIR3 T1FMY2 G7N5Q2 A0A224YPI2 A0A2K5E2G1 A0A315V885 A0A2K6S5I6 A0A0P5PVF2 A0A2I2UDS4 F7I3N3 A0A0P5DKU7 A0A2K5YWZ4 Q8NHP8 A0A2K6DCS4 A0A2K5WZV8 I2CYI6 A0A131YKS0 U3ECB3 H9YUP8 H9F441
A0A2J7PU65 J9K224 A0A2I9LPG8 A0A2H8TJI2 A0A0P5J9R6 A0A164ZX29 A0A0N8BAQ7 A0A0P6EVS2 A0A0N8B5Q0 A0A0P6GG78 A0A0P5AKM6 A0A0P5UML3 A0A0P5GTM8 A0A0N8DYW5 A0A0P5RGR3 A0A067RJD1 A0A0P5BN34 A0A0P6DC37 A0A0P5HHQ7 A0A0P6AAA5 A0A0P5QS33 A0A0P5FWY2 A0A0P6IZ23 A0A0P5IGE3 A0A0P5HKV5 A0A0P5K8J4 A0A0P5MCI9 E9GSQ2 A0A0N8E886 A0A0P5MSK5 A0A0K2SZE0 A0A0P5QAW2 A0A0P5QP37 A0A0P6FKH2 A0A1W7R9X8 R7VB97 A0A3R7PL47 A0A0P5NEB0 A0A1S3H1M5 A0A0P5JS83 A0A2P8YQZ5 T1IJU9 A0A1S3H3H8 A0A0P6ECV0 A0A0P6IGS1 A0A0P5Q498 A0A2R8QFI9 A0A0P5ZP95 A0A0P5G2M1 A0A0P5H812 A2RV19 A0A293M231 A0A3B3HSJ2 A0A0P6FAS0 V9KJN6 A0A3P9KUP8 A0A0S7G278 A0A3P9H0U9 A0A210QBW5 V9KKP6 H2LVN2 A0A3B1IZJ5 W5U8Z2 M3ZZL5 A0A0P4VWC0 V9KIX4 A0A3B4EI41 J9JWY7 V5H983 A0A034WWT4 A0A1V9XR98 A0A151N4M2 A0A060WGU2 G7PIR3 T1FMY2 G7N5Q2 A0A224YPI2 A0A2K5E2G1 A0A315V885 A0A2K6S5I6 A0A0P5PVF2 A0A2I2UDS4 F7I3N3 A0A0P5DKU7 A0A2K5YWZ4 Q8NHP8 A0A2K6DCS4 A0A2K5WZV8 I2CYI6 A0A131YKS0 U3ECB3 H9YUP8 H9F441
PDB
3FGW
E-value=1.95265e-116,
Score=1074
Ontologies
PANTHER
Topology
Subcellular location
Lysosome lumen
SignalP
Position: 1 - 16,
Likelihood: 0.950149
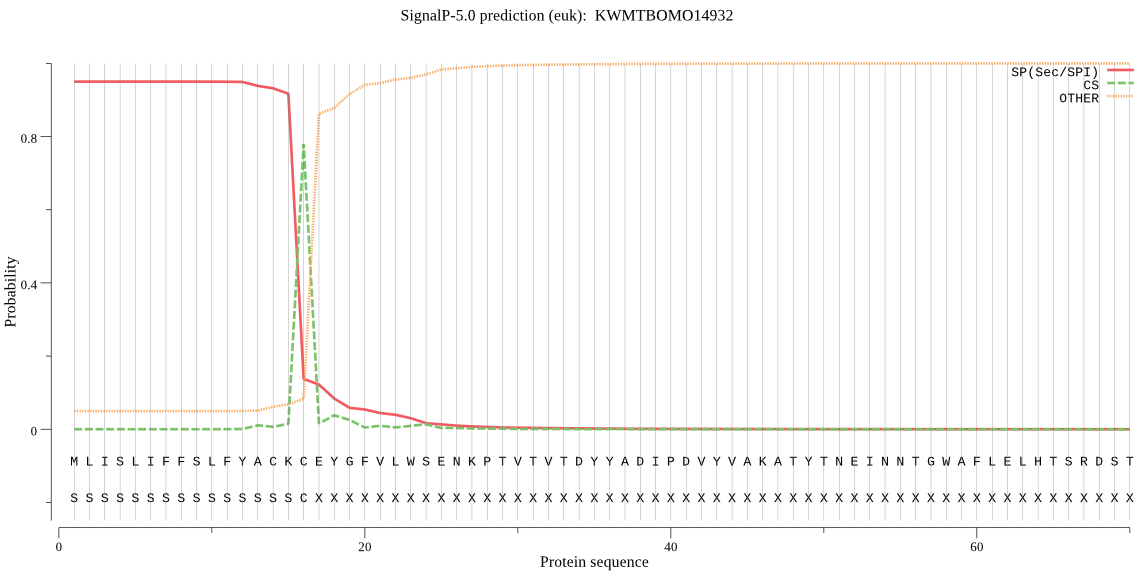
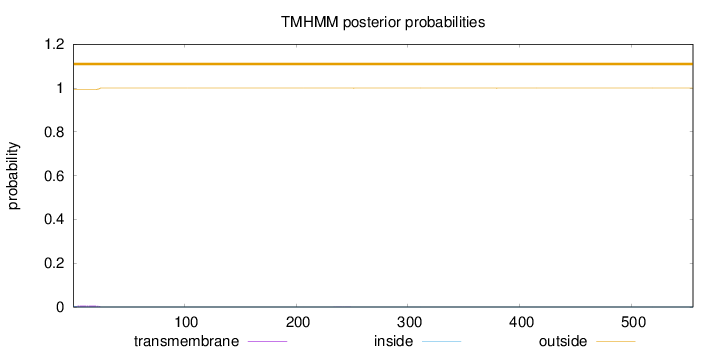
Length:
555
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.13675
Exp number, first 60 AAs:
0.13154
Total prob of N-in:
0.00618
outside
1 - 555
Population Genetic Test Statistics
Pi
252.748367
Theta
169.678585
Tajima's D
1.598757
CLR
0.059159
CSRT
0.808459577021149
Interpretation
Uncertain
