Pre Gene Modal
BGIBMGA000800
Annotation
AP-2_complex_subunit_sigma_[Solenopsis_invicta]
Full name
AP complex subunit sigma
Location in the cell
Cytoplasmic Reliability : 1.777
Sequence
CDS
ATGATAAGATTCATTTTGATCCAGAACCGAGCAGGCAAGACTCGACTTGCGAAATGGTATATGAATTTTGATGATGACGAAAAACAGAAGTTGATAGAAGAAGTGCATGCAGTCGTCACCGTCAGAGACGCGAAACACACGAACTTTGTCGAGTTCAGAAATTTCAAGATAGTATACAGGAGGTATGCAGGTCTGTACTTCTGTATATGTGTCGATGTAAATGATAACAATTTGTGTTATCTCGAGGCCATACATAACTTTGTCGAGGTCCTAAATGAATATTTCCATAATGTTTGCGAGTTGGACTTGGTCTTCAATTTTTATAAAGTGTATACGGTTGTAGACGAAATGTTTTTGGCTGGTGAGATCAGGGAGACATCCCAAACTAAAGTGCTCAAACAACTCTTGATGCTGAACTCCTTAGAATAA
Protein
MIRFILIQNRAGKTRLAKWYMNFDDDEKQKLIEEVHAVVTVRDAKHTNFVEFRNFKIVYRRYAGLYFCICVDVNDNNLCYLEAIHNFVEVLNEYFHNVCELDLVFNFYKVYTVVDEMFLAGEIRETSQTKVLKQLLMLNSLE
Summary
Similarity
Belongs to the adaptor complexes small subunit family.
Uniprot
A0A0N7Z9E3
A0A0L7L1Z4
A0A2W1BP11
D3KD02
A0A3S2M6A2
A0A139WEN5
+ More
A0A0V0GA54 A0A2A3E807 A0A2H1WME0 R4UNW1 A0A067R3E0 A0A1B6M8Z8 H9IU70 S4PDF8 A0A1B6HSN1 A0A3L8D6I2 A0A212EHB3 A0A069DW12 R4FPH9 A0A1B6FDP5 A0A0A9Z3D8 A0A1E1W7T2 A0A023F9N7 A0A224Y0K2 A0A0M8ZQE2 A0A1L8EI68 I4DP90 A0A1Q3EU05 A0A2M4CSS3 E0VYJ3 A0A194PLZ1 A0A2M4AXX4 A0A1L8DVW5 W8C7F3 D3TRZ9 A0A1I8NA29 A0A1A9YLN8 A0A0K8TYF5 A0A1A9UJU0 T1DQR4 A0A1B0BYC5 U5EV46 A0A034W3Q0 A0A182Y3R7 A0A1I8PF61 A0A0L0CJ59 A0A0A1XPQ1 A0A1L8EHV3 A0A1B0GJ46 A0A2M4C2N5 A0A1A9WY15 A0A182PSK1 A0A182MIJ9 A0A182R0Q1 A0A182SGI8 A0A182UQG5 A0A182NRV0 A0A182JRZ7 B0WQZ7 A0A182VZK8 A0A182TPD2 Q1HRK8 A0A182JDS2 A0A182WSS0 Q7PY17 A0A182F7H8 A0A182RIM5 A0A182HLM3 A0A023EET5 A0A1Y1MM25 A0A195ELI6 A0A0K8TTG1 A0A151IMN4 K7IQF3 A0A0P6IV35 A0A162NMK5 E9GIN4 A0A0P5EYG0 A0A1W4VLI3 B4MBG9 B4R122 B4KCU9 A0A3B0KW73 Q29B05 B4GZ92 T1H1D9 B3LZ31 B4NKG7 B4IKI0 B3P034 Q9VDC3 Q6XI64 A0A1B6EGN2 C4WWX9 A0A232ETT8 T1JMU4 N6TR84 A0A1W4XC34 K9IRP1 A0A1L8EHU3
A0A0V0GA54 A0A2A3E807 A0A2H1WME0 R4UNW1 A0A067R3E0 A0A1B6M8Z8 H9IU70 S4PDF8 A0A1B6HSN1 A0A3L8D6I2 A0A212EHB3 A0A069DW12 R4FPH9 A0A1B6FDP5 A0A0A9Z3D8 A0A1E1W7T2 A0A023F9N7 A0A224Y0K2 A0A0M8ZQE2 A0A1L8EI68 I4DP90 A0A1Q3EU05 A0A2M4CSS3 E0VYJ3 A0A194PLZ1 A0A2M4AXX4 A0A1L8DVW5 W8C7F3 D3TRZ9 A0A1I8NA29 A0A1A9YLN8 A0A0K8TYF5 A0A1A9UJU0 T1DQR4 A0A1B0BYC5 U5EV46 A0A034W3Q0 A0A182Y3R7 A0A1I8PF61 A0A0L0CJ59 A0A0A1XPQ1 A0A1L8EHV3 A0A1B0GJ46 A0A2M4C2N5 A0A1A9WY15 A0A182PSK1 A0A182MIJ9 A0A182R0Q1 A0A182SGI8 A0A182UQG5 A0A182NRV0 A0A182JRZ7 B0WQZ7 A0A182VZK8 A0A182TPD2 Q1HRK8 A0A182JDS2 A0A182WSS0 Q7PY17 A0A182F7H8 A0A182RIM5 A0A182HLM3 A0A023EET5 A0A1Y1MM25 A0A195ELI6 A0A0K8TTG1 A0A151IMN4 K7IQF3 A0A0P6IV35 A0A162NMK5 E9GIN4 A0A0P5EYG0 A0A1W4VLI3 B4MBG9 B4R122 B4KCU9 A0A3B0KW73 Q29B05 B4GZ92 T1H1D9 B3LZ31 B4NKG7 B4IKI0 B3P034 Q9VDC3 Q6XI64 A0A1B6EGN2 C4WWX9 A0A232ETT8 T1JMU4 N6TR84 A0A1W4XC34 K9IRP1 A0A1L8EHU3
Pubmed
27129103
26227816
28756777
18362917
19820115
24845553
+ More
19121390 23622113 30249741 22118469 26334808 25401762 26823975 25474469 22651552 20566863 26354079 24495485 20353571 25315136 25348373 25244985 26108605 25830018 17204158 17510324 12364791 14747013 17210077 24945155 26483478 28004739 26369729 20075255 21292972 17994087 15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 14525923 17550304 28648823 23537049
19121390 23622113 30249741 22118469 26334808 25401762 26823975 25474469 22651552 20566863 26354079 24495485 20353571 25315136 25348373 25244985 26108605 25830018 17204158 17510324 12364791 14747013 17210077 24945155 26483478 28004739 26369729 20075255 21292972 17994087 15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 14525923 17550304 28648823 23537049
EMBL
GDKW01000787
JAI55808.1
JTDY01003459
KOB69523.1
KZ149945
PZC76799.1
+ More
GU471224 ADC34230.1 RSAL01000022 RVE52350.1 KQ971354 KYB26396.1 GECL01001315 JAP04809.1 KZ288353 PBC27319.1 ODYU01009603 SOQ54146.1 KC741041 AGM32865.1 KK852730 KDR17519.1 GEBQ01007653 JAT32324.1 BABH01034198 GAIX01003701 JAA88859.1 GECU01030018 JAS77688.1 QOIP01000012 RLU16077.1 AGBW02014918 OWR40875.1 GBGD01003385 JAC85504.1 ACPB03003281 GAHY01000874 JAA76636.1 GECZ01030336 GECZ01021449 GECZ01003015 JAS39433.1 JAS48320.1 JAS66754.1 GBHO01003833 GBHO01003832 GDHC01022005 GDHC01019040 JAG39771.1 JAG39772.1 JAP96623.1 JAP99588.1 GDQN01008143 GDQN01004941 JAT82911.1 JAT86113.1 GBBI01000520 GEMB01000443 JAC18192.1 JAS02681.1 GFTR01002487 JAW13939.1 KQ435916 KOX68797.1 GFDG01000483 JAV18316.1 AK403474 BAM19730.1 GFDL01016245 JAV18800.1 GGFL01004199 MBW68377.1 DS235845 EEB18449.1 KQ459604 KPI92125.1 GGFK01012322 MBW45643.1 GFDF01003516 JAV10568.1 GAMC01008046 JAB98509.1 EZ424201 ADD20477.1 GDHF01033023 JAI19291.1 GAMD01002309 JAA99281.1 JXJN01022585 GANO01001183 JAB58688.1 GAKP01010544 JAC48408.1 JRES01000409 KNC31514.1 GBXI01001739 JAD12553.1 GFDG01000487 JAV18312.1 AJWK01019307 GGFJ01010439 MBW59580.1 AXCM01000082 AXCN02000250 DS232047 EDS33084.1 DQ440086 CH478277 ABF18119.1 EAT33340.1 AXCP01000903 AAAB01008987 EAA01622.2 APCN01000909 JXUM01081971 GAPW01005706 KQ563280 JAC07892.1 KXJ74134.1 GEZM01031317 JAV84936.1 KQ978730 KYN28991.1 GDAI01000378 JAI17225.1 KQ977026 KYN06218.1 GDIQ01027321 JAN67416.1 LRGB01000568 KZS18122.1 GL732546 EFX80717.1 GDIQ01265288 JAJ86436.1 CH940656 EDW58440.1 CM000364 EDX12109.1 CH933806 EDW16971.2 OUUW01000013 SPP88228.1 CM000070 EAL27194.2 CH479198 EDW28110.1 CAQQ02381001 CH902617 EDV41905.1 CH964272 EDW85139.1 CH480853 EDW51584.1 CH954181 EDV48270.1 AE014297 BT010252 KX531360 AAF55874.1 AAQ23570.1 ANY27170.1 AY231968 CM000160 AAR09991.1 EDW96244.1 GEDC01030594 GEDC01003951 GEDC01000264 JAS06704.1 JAS33347.1 JAS37034.1 ABLF02024669 AK342337 BAH72399.1 NNAY01002238 OXU21754.1 JH431868 APGK01057866 KB741282 ENN70831.1 GABZ01002828 JAA50697.1 GFDG01000512 JAV18287.1
GU471224 ADC34230.1 RSAL01000022 RVE52350.1 KQ971354 KYB26396.1 GECL01001315 JAP04809.1 KZ288353 PBC27319.1 ODYU01009603 SOQ54146.1 KC741041 AGM32865.1 KK852730 KDR17519.1 GEBQ01007653 JAT32324.1 BABH01034198 GAIX01003701 JAA88859.1 GECU01030018 JAS77688.1 QOIP01000012 RLU16077.1 AGBW02014918 OWR40875.1 GBGD01003385 JAC85504.1 ACPB03003281 GAHY01000874 JAA76636.1 GECZ01030336 GECZ01021449 GECZ01003015 JAS39433.1 JAS48320.1 JAS66754.1 GBHO01003833 GBHO01003832 GDHC01022005 GDHC01019040 JAG39771.1 JAG39772.1 JAP96623.1 JAP99588.1 GDQN01008143 GDQN01004941 JAT82911.1 JAT86113.1 GBBI01000520 GEMB01000443 JAC18192.1 JAS02681.1 GFTR01002487 JAW13939.1 KQ435916 KOX68797.1 GFDG01000483 JAV18316.1 AK403474 BAM19730.1 GFDL01016245 JAV18800.1 GGFL01004199 MBW68377.1 DS235845 EEB18449.1 KQ459604 KPI92125.1 GGFK01012322 MBW45643.1 GFDF01003516 JAV10568.1 GAMC01008046 JAB98509.1 EZ424201 ADD20477.1 GDHF01033023 JAI19291.1 GAMD01002309 JAA99281.1 JXJN01022585 GANO01001183 JAB58688.1 GAKP01010544 JAC48408.1 JRES01000409 KNC31514.1 GBXI01001739 JAD12553.1 GFDG01000487 JAV18312.1 AJWK01019307 GGFJ01010439 MBW59580.1 AXCM01000082 AXCN02000250 DS232047 EDS33084.1 DQ440086 CH478277 ABF18119.1 EAT33340.1 AXCP01000903 AAAB01008987 EAA01622.2 APCN01000909 JXUM01081971 GAPW01005706 KQ563280 JAC07892.1 KXJ74134.1 GEZM01031317 JAV84936.1 KQ978730 KYN28991.1 GDAI01000378 JAI17225.1 KQ977026 KYN06218.1 GDIQ01027321 JAN67416.1 LRGB01000568 KZS18122.1 GL732546 EFX80717.1 GDIQ01265288 JAJ86436.1 CH940656 EDW58440.1 CM000364 EDX12109.1 CH933806 EDW16971.2 OUUW01000013 SPP88228.1 CM000070 EAL27194.2 CH479198 EDW28110.1 CAQQ02381001 CH902617 EDV41905.1 CH964272 EDW85139.1 CH480853 EDW51584.1 CH954181 EDV48270.1 AE014297 BT010252 KX531360 AAF55874.1 AAQ23570.1 ANY27170.1 AY231968 CM000160 AAR09991.1 EDW96244.1 GEDC01030594 GEDC01003951 GEDC01000264 JAS06704.1 JAS33347.1 JAS37034.1 ABLF02024669 AK342337 BAH72399.1 NNAY01002238 OXU21754.1 JH431868 APGK01057866 KB741282 ENN70831.1 GABZ01002828 JAA50697.1 GFDG01000512 JAV18287.1
Proteomes
UP000037510
UP000283053
UP000007266
UP000242457
UP000027135
UP000005204
+ More
UP000279307 UP000007151 UP000015103 UP000053105 UP000009046 UP000053268 UP000095301 UP000092443 UP000078200 UP000092460 UP000076408 UP000095300 UP000037069 UP000092461 UP000091820 UP000075885 UP000075883 UP000075886 UP000075901 UP000075903 UP000075884 UP000075881 UP000002320 UP000075920 UP000075902 UP000008820 UP000075880 UP000076407 UP000007062 UP000069272 UP000075900 UP000075840 UP000069940 UP000249989 UP000078492 UP000078542 UP000002358 UP000076858 UP000000305 UP000192221 UP000008792 UP000000304 UP000009192 UP000268350 UP000001819 UP000008744 UP000015102 UP000007801 UP000007798 UP000001292 UP000008711 UP000000803 UP000002282 UP000007819 UP000215335 UP000019118 UP000192223
UP000279307 UP000007151 UP000015103 UP000053105 UP000009046 UP000053268 UP000095301 UP000092443 UP000078200 UP000092460 UP000076408 UP000095300 UP000037069 UP000092461 UP000091820 UP000075885 UP000075883 UP000075886 UP000075901 UP000075903 UP000075884 UP000075881 UP000002320 UP000075920 UP000075902 UP000008820 UP000075880 UP000076407 UP000007062 UP000069272 UP000075900 UP000075840 UP000069940 UP000249989 UP000078492 UP000078542 UP000002358 UP000076858 UP000000305 UP000192221 UP000008792 UP000000304 UP000009192 UP000268350 UP000001819 UP000008744 UP000015102 UP000007801 UP000007798 UP000001292 UP000008711 UP000000803 UP000002282 UP000007819 UP000215335 UP000019118 UP000192223
Pfam
PF01217 Clat_adaptor_s
Interpro
SUPFAM
SSF64356
SSF64356
ProteinModelPortal
A0A0N7Z9E3
A0A0L7L1Z4
A0A2W1BP11
D3KD02
A0A3S2M6A2
A0A139WEN5
+ More
A0A0V0GA54 A0A2A3E807 A0A2H1WME0 R4UNW1 A0A067R3E0 A0A1B6M8Z8 H9IU70 S4PDF8 A0A1B6HSN1 A0A3L8D6I2 A0A212EHB3 A0A069DW12 R4FPH9 A0A1B6FDP5 A0A0A9Z3D8 A0A1E1W7T2 A0A023F9N7 A0A224Y0K2 A0A0M8ZQE2 A0A1L8EI68 I4DP90 A0A1Q3EU05 A0A2M4CSS3 E0VYJ3 A0A194PLZ1 A0A2M4AXX4 A0A1L8DVW5 W8C7F3 D3TRZ9 A0A1I8NA29 A0A1A9YLN8 A0A0K8TYF5 A0A1A9UJU0 T1DQR4 A0A1B0BYC5 U5EV46 A0A034W3Q0 A0A182Y3R7 A0A1I8PF61 A0A0L0CJ59 A0A0A1XPQ1 A0A1L8EHV3 A0A1B0GJ46 A0A2M4C2N5 A0A1A9WY15 A0A182PSK1 A0A182MIJ9 A0A182R0Q1 A0A182SGI8 A0A182UQG5 A0A182NRV0 A0A182JRZ7 B0WQZ7 A0A182VZK8 A0A182TPD2 Q1HRK8 A0A182JDS2 A0A182WSS0 Q7PY17 A0A182F7H8 A0A182RIM5 A0A182HLM3 A0A023EET5 A0A1Y1MM25 A0A195ELI6 A0A0K8TTG1 A0A151IMN4 K7IQF3 A0A0P6IV35 A0A162NMK5 E9GIN4 A0A0P5EYG0 A0A1W4VLI3 B4MBG9 B4R122 B4KCU9 A0A3B0KW73 Q29B05 B4GZ92 T1H1D9 B3LZ31 B4NKG7 B4IKI0 B3P034 Q9VDC3 Q6XI64 A0A1B6EGN2 C4WWX9 A0A232ETT8 T1JMU4 N6TR84 A0A1W4XC34 K9IRP1 A0A1L8EHU3
A0A0V0GA54 A0A2A3E807 A0A2H1WME0 R4UNW1 A0A067R3E0 A0A1B6M8Z8 H9IU70 S4PDF8 A0A1B6HSN1 A0A3L8D6I2 A0A212EHB3 A0A069DW12 R4FPH9 A0A1B6FDP5 A0A0A9Z3D8 A0A1E1W7T2 A0A023F9N7 A0A224Y0K2 A0A0M8ZQE2 A0A1L8EI68 I4DP90 A0A1Q3EU05 A0A2M4CSS3 E0VYJ3 A0A194PLZ1 A0A2M4AXX4 A0A1L8DVW5 W8C7F3 D3TRZ9 A0A1I8NA29 A0A1A9YLN8 A0A0K8TYF5 A0A1A9UJU0 T1DQR4 A0A1B0BYC5 U5EV46 A0A034W3Q0 A0A182Y3R7 A0A1I8PF61 A0A0L0CJ59 A0A0A1XPQ1 A0A1L8EHV3 A0A1B0GJ46 A0A2M4C2N5 A0A1A9WY15 A0A182PSK1 A0A182MIJ9 A0A182R0Q1 A0A182SGI8 A0A182UQG5 A0A182NRV0 A0A182JRZ7 B0WQZ7 A0A182VZK8 A0A182TPD2 Q1HRK8 A0A182JDS2 A0A182WSS0 Q7PY17 A0A182F7H8 A0A182RIM5 A0A182HLM3 A0A023EET5 A0A1Y1MM25 A0A195ELI6 A0A0K8TTG1 A0A151IMN4 K7IQF3 A0A0P6IV35 A0A162NMK5 E9GIN4 A0A0P5EYG0 A0A1W4VLI3 B4MBG9 B4R122 B4KCU9 A0A3B0KW73 Q29B05 B4GZ92 T1H1D9 B3LZ31 B4NKG7 B4IKI0 B3P034 Q9VDC3 Q6XI64 A0A1B6EGN2 C4WWX9 A0A232ETT8 T1JMU4 N6TR84 A0A1W4XC34 K9IRP1 A0A1L8EHU3
PDB
4UQI
E-value=1.34749e-67,
Score=644
Ontologies
GO
PANTHER
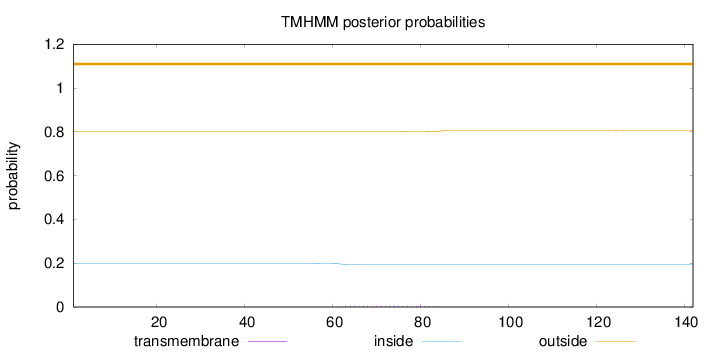
Topology
Length:
142
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.12991
Exp number, first 60 AAs:
0.00228
Total prob of N-in:
0.19955
outside
1 - 142
Population Genetic Test Statistics
Pi
171.256439
Theta
150.539161
Tajima's D
0.432624
CLR
0.778442
CSRT
0.501174941252937
Interpretation
Uncertain
