Gene
KWMTBOMO14927 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA000798
Annotation
PREDICTED:_RNA-binding_protein_Nova-2_isoform_X3_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.04 Mitochondrial Reliability : 1.108
Sequence
CDS
ATGGAACGCGCTTGTCTGATCACCGGCTCGGTGGAGGGGATCATGGTTGTGTTGGACTTCATTATGGAGAAGATTAAAGAGAAGCCAGAGCTCGTGAAGCCATTCCCGGAGGGAGTCGACACTAAAATGCCCCAGGACAGAGACAAACAGGTGAAGATCTTGGTGCCAAACTCGACAGCTGGCATGATAATCGGCAAAGGCGGCAACTACATCAAGCAGATCAAGGAACAGAGCGGCAGCTACGTTCAGATCTCGCAGAAGGCCAAGGAATTGTCCTTGCAGGAGAGGTGCATTACTGTTGTGGGCGAGAAGGAAAGCAACAAGAAGGCCTGCCTCATGATCCTTCAGAAGGTGGTTGATGACCCTCAGTCCGGGTCGTGCCCCAACGTATCATACGCGGACGTGGCCGGACCGGTCGCCAACTACAATCCGACGGGATCGCCCTACGCTGTACCCACCGCCGAGAGTCACGGGCTCGGCGGTGCGGTGGGCGGAGTGGGCGGGGTGCTGATGAACGGCGGCGGCGCGCTGGGCGGGCTGGGCGCGCTCGCCCTGCAGCTGTACCTGGCGCCGCCCGGCACCCCGCCCTCCGCGCCCATCACGCAGCACACCCTCGACCACATCAAGGGCGCGCTGCGGCAGGCCGGCTACAGCGAGGCTGGGGTGAGCGAGATCGGCGCGGCGCTGGGGCTGCTGGTGAAGCACGGCGTGCTGGGGCTGGGCGTGGGGGGCCCGATGCCCGCGCCGCCCGCGCTCTCCGCCGCCTACTTCCCACTGCCCGCGCAGGACCCGCCCGCCGTCTTCGGACCCCTCGCGCAGGTCTCGCTAGGCGGCGCGCGCGGCGGCTCGCTCGACCGTTTCGCGGAGGTTGCATTCGAGGCGCTTCGCCCCCCAGCCGTGGCGCCCATCTCGCTGCAAGGCGGCGTGCCGTTCCCGTCGGCCTCGCTGCTGCCGCTGTCCAAGAGCCCCACGCCGGCCGACGCGCAGCCCAAGGACTCTAAGAACGTCGAGATAGCGGAGGTCATCGTCGGCGCCATCCTCGGGCCGGGAGGGCGCAGCCTGGTGGAGATACAGCAGATGTCCGGGGCCAACATCCAGATCTCGAAGAAGGGCACGTTCGCTCCCGGCACCCGGAACCGCATCGTCACCATCTCCGGCTCGCAGACGGCCATCAGCAACGCTCACTACCTCATCGAGCAGAAGATCCAGGAGGAGGAGCTGAAGCGGACCAGGCACAACGCCATCTCGACCCTCATGCAGTAA
Protein
MERACLITGSVEGIMVVLDFIMEKIKEKPELVKPFPEGVDTKMPQDRDKQVKILVPNSTAGMIIGKGGNYIKQIKEQSGSYVQISQKAKELSLQERCITVVGEKESNKKACLMILQKVVDDPQSGSCPNVSYADVAGPVANYNPTGSPYAVPTAESHGLGGAVGGVGGVLMNGGGALGGLGALALQLYLAPPGTPPSAPITQHTLDHIKGALRQAGYSEAGVSEIGAALGLLVKHGVLGLGVGGPMPAPPALSAAYFPLPAQDPPAVFGPLAQVSLGGARGGSLDRFAEVAFEALRPPAVAPISLQGGVPFPSASLLPLSKSPTPADAQPKDSKNVEIAEVIVGAILGPGGRSLVEIQQMSGANIQISKKGTFAPGTRNRIVTISGSQTAISNAHYLIEQKIQEEELKRTRHNAISTLMQ
Summary
Uniprot
A0A194RHY9
A0A212EHC0
A0A194PFI4
A0A0L7LG18
A0A2H1VS90
A0A1Y1LIP9
+ More
A0A1Y1LH63 V5GMR4 D6WPZ1 U4UI29 N6UC93 A0A2P8XX37 E2B6J6 A0A026WT92 A0A2A3ERT9 V9II74 A0A088AB41 A0A0J7KEI4 A0A182GXK6 A0A2M4A663 E9JAZ8 A0A195BH93 A0A195C006 A0A2M4A5S1 F4WFW8 A0A151JSH0 A0A195F1N4 E2ABZ7 A0A151XBI4 A0A3L8DAM9 Q16EY8 A0A182FAC2 A0A1L8DH20 A0A158NLA9 A0A1L8DG89 A0A023ESU8 T1DI40 A0A1B6ED52 A0A1B6DRI1 A0A1S4GG92 A0A2C9GQG9 Q7QD06 A0A1Y9H210 A0A182XHJ2 A0A182UTK8 A0A1B6EHS5 A0A2C9GXC1 A0A2C9GQA4 A0A0B4KFY5 A0A1B6M1I5 Q8T4D1 A0A0R1E1V8 A0A2C9GQA0 Q0KI96 A0A2C9GXB5 A0A182VRM5 Q9GQN4 Q9GQN3 B4HKD5 Q8INP0 A0A0B4LGY5 Q8IGS7 A0A182XV56 A0A0B4LI06 A0A023UKB8 A0A1Y9H2H6 A0A182P630 A0A182MM01 B4JEK8 A0A0B4JCX5 A0A0B4KGY6 A0A0R1E1W7 A0A0R1E6F8 A0A0R1E1W9 A0A0R1E6D5 A0A0R1E299 A0A1W4W2N2 A0A1W4VPY2 A0A0R1E2F8 A0A0R1E6D8 B4PUN7 B3NZS2 A0A0R1E3B8 A0A1W4W2M2 A0A023F1K7 A0A224XQ52 B7PQM3 A0A1W4VPL9 A0A1W4VPW7 A0A0K8T1F7 A0A0A9Y503 A0A2M4A7C4 W5J335 A0A2M3Z1X7 A0A0P6EHZ8 A0A2M4CH96 A0A2M4CGZ4
A0A1Y1LH63 V5GMR4 D6WPZ1 U4UI29 N6UC93 A0A2P8XX37 E2B6J6 A0A026WT92 A0A2A3ERT9 V9II74 A0A088AB41 A0A0J7KEI4 A0A182GXK6 A0A2M4A663 E9JAZ8 A0A195BH93 A0A195C006 A0A2M4A5S1 F4WFW8 A0A151JSH0 A0A195F1N4 E2ABZ7 A0A151XBI4 A0A3L8DAM9 Q16EY8 A0A182FAC2 A0A1L8DH20 A0A158NLA9 A0A1L8DG89 A0A023ESU8 T1DI40 A0A1B6ED52 A0A1B6DRI1 A0A1S4GG92 A0A2C9GQG9 Q7QD06 A0A1Y9H210 A0A182XHJ2 A0A182UTK8 A0A1B6EHS5 A0A2C9GXC1 A0A2C9GQA4 A0A0B4KFY5 A0A1B6M1I5 Q8T4D1 A0A0R1E1V8 A0A2C9GQA0 Q0KI96 A0A2C9GXB5 A0A182VRM5 Q9GQN4 Q9GQN3 B4HKD5 Q8INP0 A0A0B4LGY5 Q8IGS7 A0A182XV56 A0A0B4LI06 A0A023UKB8 A0A1Y9H2H6 A0A182P630 A0A182MM01 B4JEK8 A0A0B4JCX5 A0A0B4KGY6 A0A0R1E1W7 A0A0R1E6F8 A0A0R1E1W9 A0A0R1E6D5 A0A0R1E299 A0A1W4W2N2 A0A1W4VPY2 A0A0R1E2F8 A0A0R1E6D8 B4PUN7 B3NZS2 A0A0R1E3B8 A0A1W4W2M2 A0A023F1K7 A0A224XQ52 B7PQM3 A0A1W4VPL9 A0A1W4VPW7 A0A0K8T1F7 A0A0A9Y503 A0A2M4A7C4 W5J335 A0A2M3Z1X7 A0A0P6EHZ8 A0A2M4CH96 A0A2M4CGZ4
Pubmed
26354079
22118469
26227816
28004739
18362917
19820115
+ More
23537049 29403074 20798317 24508170 26483478 21282665 21719571 30249741 17510324 21347285 24945155 24330624 12364791 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17994087 17550304 25244985 25474469 25401762 26823975 20920257 23761445
23537049 29403074 20798317 24508170 26483478 21282665 21719571 30249741 17510324 21347285 24945155 24330624 12364791 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17994087 17550304 25244985 25474469 25401762 26823975 20920257 23761445
EMBL
KQ460205
KPJ17054.1
AGBW02014918
OWR40870.1
KQ459604
KPI92131.1
+ More
JTDY01001252 KOB74397.1 ODYU01004145 SOQ43705.1 GEZM01057603 GEZM01057601 JAV72230.1 GEZM01057604 GEZM01057602 JAV72231.1 GALX01003077 JAB65389.1 KQ971354 EFA06153.2 KB632333 ERL92682.1 APGK01035299 KB740923 ENN78251.1 PYGN01001220 PSN36570.1 GL445975 EFN88688.1 KK107111 EZA58891.1 KZ288194 PBC33839.1 JR048992 AEY60828.1 LBMM01008781 KMQ88644.1 JXUM01095512 KQ564200 KXJ72596.1 GGFK01002797 MBW36118.1 GL770707 EFZ09994.1 KQ976488 KYM83524.1 KQ978501 KYM93506.1 GGFK01002657 MBW35978.1 GL888128 EGI66735.1 KQ978530 KYN30319.1 KQ981866 KYN34082.1 GL438389 EFN69015.1 KQ982320 KYQ57732.1 QOIP01000011 RLU16978.1 CH478548 EAT32800.1 GFDF01008332 JAV05752.1 ADTU01019250 ADTU01019251 ADTU01019252 ADTU01019253 ADTU01019254 GFDF01008730 JAV05354.1 GAPW01001236 JAC12362.1 GALA01001166 JAA93686.1 GEDC01001432 JAS35866.1 GEDC01009004 JAS28294.1 AAAB01008859 APCN01000586 EAA08028.5 GECZ01032290 JAS37479.1 AE014297 AGB95796.1 GEBQ01010195 JAT29782.1 AY089247 AAL89985.1 ABI31154.1 CM000160 KRK03260.1 ABI31152.1 ADV37292.1 ADV37293.1 AF220422 AAF54377.3 AAG36789.1 AAN13424.2 AAN13425.2 ADV37290.1 ADV37294.1 AF220423 AAF54378.2 AAG36790.1 CH480815 EDW42885.1 AAN13428.1 AHN57251.1 BT001623 AAN71378.1 AHN57252.1 BT150444 AHY03616.1 AXCM01000411 AXCM01000412 CH916369 EDV93139.1 ADV37291.1 AGB95797.1 KRK03259.1 KRK03268.1 KRK03261.1 KRK03269.1 KRK03258.1 KRK03257.1 KRK03263.1 KRK03264.1 KRK03271.1 KRK03266.1 EDW96654.1 KRK03265.1 KRK03267.1 KRK03270.1 CH954181 EDV49782.1 KRK03262.1 GBBI01003529 JAC15183.1 GFTR01006295 JAW10131.1 ABJB010402307 DS766405 EEC08895.1 GBRD01006821 JAG59000.1 GBHO01031266 GBHO01015482 GDHC01001637 JAG12338.1 JAG28122.1 JAQ16992.1 GGFK01003309 MBW36630.1 ADMH02002218 ETN57733.1 GGFM01001786 MBW22537.1 GDIQ01075248 JAN19489.1 GGFL01000451 MBW64629.1 GGFL01000442 MBW64620.1
JTDY01001252 KOB74397.1 ODYU01004145 SOQ43705.1 GEZM01057603 GEZM01057601 JAV72230.1 GEZM01057604 GEZM01057602 JAV72231.1 GALX01003077 JAB65389.1 KQ971354 EFA06153.2 KB632333 ERL92682.1 APGK01035299 KB740923 ENN78251.1 PYGN01001220 PSN36570.1 GL445975 EFN88688.1 KK107111 EZA58891.1 KZ288194 PBC33839.1 JR048992 AEY60828.1 LBMM01008781 KMQ88644.1 JXUM01095512 KQ564200 KXJ72596.1 GGFK01002797 MBW36118.1 GL770707 EFZ09994.1 KQ976488 KYM83524.1 KQ978501 KYM93506.1 GGFK01002657 MBW35978.1 GL888128 EGI66735.1 KQ978530 KYN30319.1 KQ981866 KYN34082.1 GL438389 EFN69015.1 KQ982320 KYQ57732.1 QOIP01000011 RLU16978.1 CH478548 EAT32800.1 GFDF01008332 JAV05752.1 ADTU01019250 ADTU01019251 ADTU01019252 ADTU01019253 ADTU01019254 GFDF01008730 JAV05354.1 GAPW01001236 JAC12362.1 GALA01001166 JAA93686.1 GEDC01001432 JAS35866.1 GEDC01009004 JAS28294.1 AAAB01008859 APCN01000586 EAA08028.5 GECZ01032290 JAS37479.1 AE014297 AGB95796.1 GEBQ01010195 JAT29782.1 AY089247 AAL89985.1 ABI31154.1 CM000160 KRK03260.1 ABI31152.1 ADV37292.1 ADV37293.1 AF220422 AAF54377.3 AAG36789.1 AAN13424.2 AAN13425.2 ADV37290.1 ADV37294.1 AF220423 AAF54378.2 AAG36790.1 CH480815 EDW42885.1 AAN13428.1 AHN57251.1 BT001623 AAN71378.1 AHN57252.1 BT150444 AHY03616.1 AXCM01000411 AXCM01000412 CH916369 EDV93139.1 ADV37291.1 AGB95797.1 KRK03259.1 KRK03268.1 KRK03261.1 KRK03269.1 KRK03258.1 KRK03257.1 KRK03263.1 KRK03264.1 KRK03271.1 KRK03266.1 EDW96654.1 KRK03265.1 KRK03267.1 KRK03270.1 CH954181 EDV49782.1 KRK03262.1 GBBI01003529 JAC15183.1 GFTR01006295 JAW10131.1 ABJB010402307 DS766405 EEC08895.1 GBRD01006821 JAG59000.1 GBHO01031266 GBHO01015482 GDHC01001637 JAG12338.1 JAG28122.1 JAQ16992.1 GGFK01003309 MBW36630.1 ADMH02002218 ETN57733.1 GGFM01001786 MBW22537.1 GDIQ01075248 JAN19489.1 GGFL01000451 MBW64629.1 GGFL01000442 MBW64620.1
Proteomes
UP000053240
UP000007151
UP000053268
UP000037510
UP000007266
UP000030742
+ More
UP000019118 UP000245037 UP000008237 UP000053097 UP000242457 UP000005203 UP000036403 UP000069940 UP000249989 UP000078540 UP000078542 UP000007755 UP000078492 UP000078541 UP000000311 UP000075809 UP000279307 UP000008820 UP000069272 UP000005205 UP000075840 UP000007062 UP000075884 UP000076407 UP000075903 UP000000803 UP000002282 UP000075920 UP000001292 UP000076408 UP000075885 UP000075883 UP000001070 UP000192221 UP000008711 UP000001555 UP000000673
UP000019118 UP000245037 UP000008237 UP000053097 UP000242457 UP000005203 UP000036403 UP000069940 UP000249989 UP000078540 UP000078542 UP000007755 UP000078492 UP000078541 UP000000311 UP000075809 UP000279307 UP000008820 UP000069272 UP000005205 UP000075840 UP000007062 UP000075884 UP000076407 UP000075903 UP000000803 UP000002282 UP000075920 UP000001292 UP000076408 UP000075885 UP000075883 UP000001070 UP000192221 UP000008711 UP000001555 UP000000673
Pfam
PF00013 KH_1
SUPFAM
SSF54791
SSF54791
Gene 3D
ProteinModelPortal
A0A194RHY9
A0A212EHC0
A0A194PFI4
A0A0L7LG18
A0A2H1VS90
A0A1Y1LIP9
+ More
A0A1Y1LH63 V5GMR4 D6WPZ1 U4UI29 N6UC93 A0A2P8XX37 E2B6J6 A0A026WT92 A0A2A3ERT9 V9II74 A0A088AB41 A0A0J7KEI4 A0A182GXK6 A0A2M4A663 E9JAZ8 A0A195BH93 A0A195C006 A0A2M4A5S1 F4WFW8 A0A151JSH0 A0A195F1N4 E2ABZ7 A0A151XBI4 A0A3L8DAM9 Q16EY8 A0A182FAC2 A0A1L8DH20 A0A158NLA9 A0A1L8DG89 A0A023ESU8 T1DI40 A0A1B6ED52 A0A1B6DRI1 A0A1S4GG92 A0A2C9GQG9 Q7QD06 A0A1Y9H210 A0A182XHJ2 A0A182UTK8 A0A1B6EHS5 A0A2C9GXC1 A0A2C9GQA4 A0A0B4KFY5 A0A1B6M1I5 Q8T4D1 A0A0R1E1V8 A0A2C9GQA0 Q0KI96 A0A2C9GXB5 A0A182VRM5 Q9GQN4 Q9GQN3 B4HKD5 Q8INP0 A0A0B4LGY5 Q8IGS7 A0A182XV56 A0A0B4LI06 A0A023UKB8 A0A1Y9H2H6 A0A182P630 A0A182MM01 B4JEK8 A0A0B4JCX5 A0A0B4KGY6 A0A0R1E1W7 A0A0R1E6F8 A0A0R1E1W9 A0A0R1E6D5 A0A0R1E299 A0A1W4W2N2 A0A1W4VPY2 A0A0R1E2F8 A0A0R1E6D8 B4PUN7 B3NZS2 A0A0R1E3B8 A0A1W4W2M2 A0A023F1K7 A0A224XQ52 B7PQM3 A0A1W4VPL9 A0A1W4VPW7 A0A0K8T1F7 A0A0A9Y503 A0A2M4A7C4 W5J335 A0A2M3Z1X7 A0A0P6EHZ8 A0A2M4CH96 A0A2M4CGZ4
A0A1Y1LH63 V5GMR4 D6WPZ1 U4UI29 N6UC93 A0A2P8XX37 E2B6J6 A0A026WT92 A0A2A3ERT9 V9II74 A0A088AB41 A0A0J7KEI4 A0A182GXK6 A0A2M4A663 E9JAZ8 A0A195BH93 A0A195C006 A0A2M4A5S1 F4WFW8 A0A151JSH0 A0A195F1N4 E2ABZ7 A0A151XBI4 A0A3L8DAM9 Q16EY8 A0A182FAC2 A0A1L8DH20 A0A158NLA9 A0A1L8DG89 A0A023ESU8 T1DI40 A0A1B6ED52 A0A1B6DRI1 A0A1S4GG92 A0A2C9GQG9 Q7QD06 A0A1Y9H210 A0A182XHJ2 A0A182UTK8 A0A1B6EHS5 A0A2C9GXC1 A0A2C9GQA4 A0A0B4KFY5 A0A1B6M1I5 Q8T4D1 A0A0R1E1V8 A0A2C9GQA0 Q0KI96 A0A2C9GXB5 A0A182VRM5 Q9GQN4 Q9GQN3 B4HKD5 Q8INP0 A0A0B4LGY5 Q8IGS7 A0A182XV56 A0A0B4LI06 A0A023UKB8 A0A1Y9H2H6 A0A182P630 A0A182MM01 B4JEK8 A0A0B4JCX5 A0A0B4KGY6 A0A0R1E1W7 A0A0R1E6F8 A0A0R1E1W9 A0A0R1E6D5 A0A0R1E299 A0A1W4W2N2 A0A1W4VPY2 A0A0R1E2F8 A0A0R1E6D8 B4PUN7 B3NZS2 A0A0R1E3B8 A0A1W4W2M2 A0A023F1K7 A0A224XQ52 B7PQM3 A0A1W4VPL9 A0A1W4VPW7 A0A0K8T1F7 A0A0A9Y503 A0A2M4A7C4 W5J335 A0A2M3Z1X7 A0A0P6EHZ8 A0A2M4CH96 A0A2M4CGZ4
PDB
2ANR
E-value=1.69465e-24,
Score=279
Ontologies
GO
PANTHER
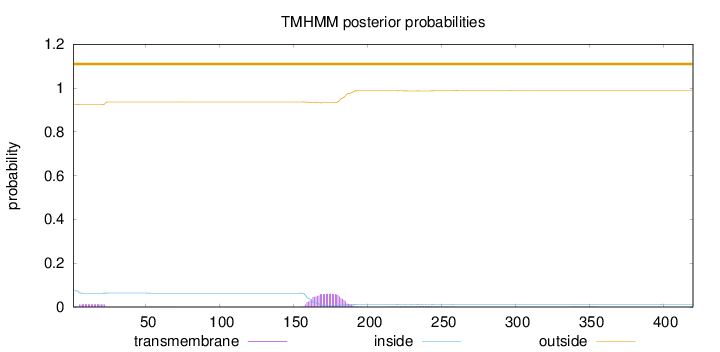
Topology
Length:
420
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.68451
Exp number, first 60 AAs:
0.22221
Total prob of N-in:
0.07416
outside
1 - 420
Population Genetic Test Statistics
Pi
221.331448
Theta
186.681759
Tajima's D
0.418185
CLR
0.631191
CSRT
0.499975001249937
Interpretation
Uncertain
