Pre Gene Modal
BGIBMGA000793
Annotation
PREDICTED:_protein_YIF1A_isoform_X1_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 3.159
Sequence
CDS
ATGAATTTTAACGCGTCCAGAAACGTAGGGCCTTCAGGTGCCATCCGTAAAGCAAAACGAGTAAGTGATGTAGCTGCCATGGGTACTCCAGCACCTAACCCTGCATTTAGTCCCACTGCATATAGCCCTGGGTATGCACCTACAGGACCTCCTCCGCCTTACCAGGAAAGCTTCCAATTAGATGCCAATCAAGATTTTAATACACATGCTCCTGCAGGTAATTTTGGATTTGCTGGGGGATTTCCTTCTCCAATGACTTCCCCGGCGCAAATAAGCTCTATGCTGCAACAACCTGTAGTCCAGGACATGGCTATTCAATACGGCAACCAGCTCGCGGCTCAAGGCAAGGAGGCTGTGCAACGAGAATTGCACAAGTTTGTGCCAGTCTCGAGGCTGCGGTACTATTTTGCAGTCGATACAAGATATGTTATAAGGAAACTAATGCTCATAGTCTTCCCCTATACTCATAAGGAGTGGATGGTGAAATACGATCAAGACACTCCAGTGCAGCCCCGGTACGACATCAACGCCCCGGACCTGTACATACCCTCCATGGGGTACGTCACGTACGTGCTGCTGGCCGGCTTCATGTTAGGTCTTCAGCACAGATTCTCACCGGAACAAATAGGCATCCAGGCGTCCAGTGCCCTGGCCTACATTATCTTCGAGATGCTGCTGTACCTCGTCACGCTGTACGTCACCAACACCAGCACCAGCCTCAAGACCCTGGACCTGCTGGCGTACTCCGGCTACAAGTACACCGTGATGATCTCCAGCCTGCTGGCCGGCCTGCTCGCGGGCAGGACGGGCTACTACTGCGGCCTGCTCTACAGCAGCTGCGCTTTGTCTTACTTTTTGGTGAAGACGCTTCGTCTCCAGTTGCTGTCGGGGTCTCAGGGTCCGGAGCAGCCCTCGTACGGGTTCCCCAACCCTCCCTACGCCGCCAACCCTTACTCCGACGCCTGGGACAAACCAACACCAGGAGGCACCAAGCGAAGGGTCTACTTCTTGCTGTTCGTGGCGATAACACAGCCGCTGCTGTGCTGGTGGCTGACTTACCATCTCGTGGCGTCGCCGCCTGGCGAGGGCGCGGGCCCGGCGCGATAG
Protein
MNFNASRNVGPSGAIRKAKRVSDVAAMGTPAPNPAFSPTAYSPGYAPTGPPPPYQESFQLDANQDFNTHAPAGNFGFAGGFPSPMTSPAQISSMLQQPVVQDMAIQYGNQLAAQGKEAVQRELHKFVPVSRLRYYFAVDTRYVIRKLMLIVFPYTHKEWMVKYDQDTPVQPRYDINAPDLYIPSMGYVTYVLLAGFMLGLQHRFSPEQIGIQASSALAYIIFEMLLYLVTLYVTNTSTSLKTLDLLAYSGYKYTVMISSLLAGLLAGRTGYYCGLLYSSCALSYFLVKTLRLQLLSGSQGPEQPSYGFPNPPYAANPYSDAWDKPTPGGTKRRVYFLLFVAITQPLLCWWLTYHLVASPPGEGAGPAR
Summary
Uniprot
H9IU63
A0A194RHL2
A0A2H1W5X4
A0A194PHE7
A0A3S2NKD0
A0A212EHB2
+ More
A0A1E1WQL9 A0A2W1BYE8 I4DP89 D6WTI1 V5GPC0 A0A1L8D967 A0A1L8D949 N6TID4 A0A067RD41 A0A1Q3FBX3 U5EVY5 A0A1W4XRD5 A0A1B0CS20 A0A023EPS3 B0XHY4 Q1HQJ2 A0A182MN20 T1EAH5 A0A182VYE2 A0A182XWI1 A0A182LQQ1 A0A2J7QBS7 A0A2J7QBT5 T1DRV1 A0A2J7QBS0 A0A336MTE9 A0A182FQN7 A0A182WZI9 Q7QK67 A0A0K8U137 A0A0K8W6F8 A0A2M4AI20 A0A2M4BR45 A0A0K8TMF3 A0A0A1XN97 W5J4W1 A0A0A1WEZ0 A0A182TD98 A0A034WEU8 A0A034WGE6 A0A182HZN8 A0A1A9XDJ3 A0A1B0C4H0 A0A1A9ZGF1 W8BUF6 W8C161 A0A182PEB3 A0A182IM96 A0A2M3Z6T9 A0A2M3ZHY3 D3TMP1 Q8MTA2 Q8IMR3 A0A336MJU1 H8F4V3 Q6NNZ6 Q9VBC5 Q8IMR4 A0A1A9WSG8 A0A336MM83 A0A182Q771 A0A182NMW1 A0A0L0CQF8 A0A1B6E7H8 B4NKN4 A0A1Y1L7J5 A0A1Y1L7H1 A0A336MDY0 A0A0M4EJ72 A0A1I8Q7P3 T1JB28 A0A1I8Q7S5 A0A0Q9WTC3 A0A0P4WE48 B4LWB2 A0A0Q9WHG8 T1PCH3 A0A0P9CAS5 A0A1I8MJV6 A0A1B6IJP9 A0A0Q9X2V1 B3M3C8 A0A0Q9X2G0 B4K8Y8 A0A0P8YNK6 A0A1I8MJW2 A0A1W4VM29 A0A1W4VLC4 A0A1W4V8Y5 A0A3B0JL82 A0A3B0K004 B4JY65 A0A3B0K7W4 A0A0R1E554
A0A1E1WQL9 A0A2W1BYE8 I4DP89 D6WTI1 V5GPC0 A0A1L8D967 A0A1L8D949 N6TID4 A0A067RD41 A0A1Q3FBX3 U5EVY5 A0A1W4XRD5 A0A1B0CS20 A0A023EPS3 B0XHY4 Q1HQJ2 A0A182MN20 T1EAH5 A0A182VYE2 A0A182XWI1 A0A182LQQ1 A0A2J7QBS7 A0A2J7QBT5 T1DRV1 A0A2J7QBS0 A0A336MTE9 A0A182FQN7 A0A182WZI9 Q7QK67 A0A0K8U137 A0A0K8W6F8 A0A2M4AI20 A0A2M4BR45 A0A0K8TMF3 A0A0A1XN97 W5J4W1 A0A0A1WEZ0 A0A182TD98 A0A034WEU8 A0A034WGE6 A0A182HZN8 A0A1A9XDJ3 A0A1B0C4H0 A0A1A9ZGF1 W8BUF6 W8C161 A0A182PEB3 A0A182IM96 A0A2M3Z6T9 A0A2M3ZHY3 D3TMP1 Q8MTA2 Q8IMR3 A0A336MJU1 H8F4V3 Q6NNZ6 Q9VBC5 Q8IMR4 A0A1A9WSG8 A0A336MM83 A0A182Q771 A0A182NMW1 A0A0L0CQF8 A0A1B6E7H8 B4NKN4 A0A1Y1L7J5 A0A1Y1L7H1 A0A336MDY0 A0A0M4EJ72 A0A1I8Q7P3 T1JB28 A0A1I8Q7S5 A0A0Q9WTC3 A0A0P4WE48 B4LWB2 A0A0Q9WHG8 T1PCH3 A0A0P9CAS5 A0A1I8MJV6 A0A1B6IJP9 A0A0Q9X2V1 B3M3C8 A0A0Q9X2G0 B4K8Y8 A0A0P8YNK6 A0A1I8MJW2 A0A1W4VM29 A0A1W4VLC4 A0A1W4V8Y5 A0A3B0JL82 A0A3B0K004 B4JY65 A0A3B0K7W4 A0A0R1E554
Pubmed
19121390
26354079
22118469
28756777
22651552
18362917
+ More
19820115 23537049 24845553 24945155 17204158 17510324 25244985 20966253 12364791 14747013 17210077 26369729 25830018 20920257 23761445 25348373 24495485 20353571 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 26108605 17994087 28004739 25315136 17550304
19820115 23537049 24845553 24945155 17204158 17510324 25244985 20966253 12364791 14747013 17210077 26369729 25830018 20920257 23761445 25348373 24495485 20353571 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 26108605 17994087 28004739 25315136 17550304
EMBL
BABH01034187
KQ460205
KPJ17057.1
ODYU01006241
SOQ47904.1
KQ459604
+ More
KPI92134.1 RSAL01000057 RVE49782.1 AGBW02014918 OWR40868.1 GDQN01001754 JAT89300.1 KZ149945 PZC76793.1 AK403473 BAM19729.1 KQ971354 EFA07180.1 GALX01002512 JAB65954.1 GFDF01011086 JAV02998.1 GFDF01011085 JAV02999.1 APGK01025209 KB740577 KB631756 ENN80184.1 ERL85820.1 KK852577 KDR20942.1 GFDL01009988 JAV25057.1 GANO01003201 JAB56670.1 AJWK01025575 GAPW01002071 JAC11527.1 DS233230 EDS28830.1 DQ440452 CH477345 ABF18485.1 EAT43008.1 AXCM01004865 GAMD01000407 JAB01184.1 NEVH01016296 PNF26032.1 PNF26035.1 GAMD01000490 JAB01101.1 PNF26034.1 UFQT01001170 SSX29248.1 AAAB01008799 EAA03725.5 GDHF01031855 JAI20459.1 GDHF01005556 JAI46758.1 GGFK01006937 MBW40258.1 GGFJ01006351 MBW55492.1 GDAI01002056 JAI15547.1 GBXI01001796 JAD12496.1 ADMH02002134 ETN58433.1 GBXI01016718 JAC97573.1 GAKP01006110 JAC52842.1 GAKP01006109 JAC52843.1 APCN01001988 JXJN01025444 GAMC01003713 JAC02843.1 GAMC01003712 JAC02844.1 GGFM01003463 MBW24214.1 GGFM01007287 MBW28038.1 EZ422693 ADD18969.1 AY118289 AAM48318.1 AE014297 BT044376 AAN14084.1 ACH92441.1 SSX29249.1 BT133343 AFF18559.1 BT011135 AAR82802.1 BT133010 AAF56617.1 AEX66196.1 AAN14083.1 UFQT01000773 SSX27128.1 AXCN02001551 JRES01000062 KNC34476.1 GEDC01003406 JAS33892.1 CH964272 EDW84095.2 GEZM01062522 GEZM01062515 JAV69663.1 GEZM01062526 GEZM01062517 JAV69629.1 SSX27129.1 CP012526 ALC46896.1 JH432008 CH940650 KRF83381.1 GDRN01065133 JAI64729.1 EDW67646.1 KRF83382.1 KA645643 AFP60272.1 CH902617 KPU80345.1 GECU01020560 JAS87146.1 CH933806 KRG02221.1 EDV43589.1 KRG02220.1 EDW16585.1 KPU80344.1 OUUW01000005 SPP81092.1 SPP81090.1 CH916377 EDV90627.1 SPP81091.1 CM000160 KRK04374.1
KPI92134.1 RSAL01000057 RVE49782.1 AGBW02014918 OWR40868.1 GDQN01001754 JAT89300.1 KZ149945 PZC76793.1 AK403473 BAM19729.1 KQ971354 EFA07180.1 GALX01002512 JAB65954.1 GFDF01011086 JAV02998.1 GFDF01011085 JAV02999.1 APGK01025209 KB740577 KB631756 ENN80184.1 ERL85820.1 KK852577 KDR20942.1 GFDL01009988 JAV25057.1 GANO01003201 JAB56670.1 AJWK01025575 GAPW01002071 JAC11527.1 DS233230 EDS28830.1 DQ440452 CH477345 ABF18485.1 EAT43008.1 AXCM01004865 GAMD01000407 JAB01184.1 NEVH01016296 PNF26032.1 PNF26035.1 GAMD01000490 JAB01101.1 PNF26034.1 UFQT01001170 SSX29248.1 AAAB01008799 EAA03725.5 GDHF01031855 JAI20459.1 GDHF01005556 JAI46758.1 GGFK01006937 MBW40258.1 GGFJ01006351 MBW55492.1 GDAI01002056 JAI15547.1 GBXI01001796 JAD12496.1 ADMH02002134 ETN58433.1 GBXI01016718 JAC97573.1 GAKP01006110 JAC52842.1 GAKP01006109 JAC52843.1 APCN01001988 JXJN01025444 GAMC01003713 JAC02843.1 GAMC01003712 JAC02844.1 GGFM01003463 MBW24214.1 GGFM01007287 MBW28038.1 EZ422693 ADD18969.1 AY118289 AAM48318.1 AE014297 BT044376 AAN14084.1 ACH92441.1 SSX29249.1 BT133343 AFF18559.1 BT011135 AAR82802.1 BT133010 AAF56617.1 AEX66196.1 AAN14083.1 UFQT01000773 SSX27128.1 AXCN02001551 JRES01000062 KNC34476.1 GEDC01003406 JAS33892.1 CH964272 EDW84095.2 GEZM01062522 GEZM01062515 JAV69663.1 GEZM01062526 GEZM01062517 JAV69629.1 SSX27129.1 CP012526 ALC46896.1 JH432008 CH940650 KRF83381.1 GDRN01065133 JAI64729.1 EDW67646.1 KRF83382.1 KA645643 AFP60272.1 CH902617 KPU80345.1 GECU01020560 JAS87146.1 CH933806 KRG02221.1 EDV43589.1 KRG02220.1 EDW16585.1 KPU80344.1 OUUW01000005 SPP81092.1 SPP81090.1 CH916377 EDV90627.1 SPP81091.1 CM000160 KRK04374.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000283053
UP000007151
UP000007266
+ More
UP000019118 UP000030742 UP000027135 UP000192223 UP000092461 UP000002320 UP000008820 UP000075883 UP000075920 UP000076408 UP000075882 UP000235965 UP000069272 UP000076407 UP000007062 UP000000673 UP000075902 UP000075840 UP000092443 UP000092460 UP000092445 UP000075885 UP000075880 UP000000803 UP000091820 UP000075886 UP000075884 UP000037069 UP000007798 UP000092553 UP000095300 UP000008792 UP000007801 UP000095301 UP000009192 UP000192221 UP000268350 UP000001070 UP000002282
UP000019118 UP000030742 UP000027135 UP000192223 UP000092461 UP000002320 UP000008820 UP000075883 UP000075920 UP000076408 UP000075882 UP000235965 UP000069272 UP000076407 UP000007062 UP000000673 UP000075902 UP000075840 UP000092443 UP000092460 UP000092445 UP000075885 UP000075880 UP000000803 UP000091820 UP000075886 UP000075884 UP000037069 UP000007798 UP000092553 UP000095300 UP000008792 UP000007801 UP000095301 UP000009192 UP000192221 UP000268350 UP000001070 UP000002282
ProteinModelPortal
H9IU63
A0A194RHL2
A0A2H1W5X4
A0A194PHE7
A0A3S2NKD0
A0A212EHB2
+ More
A0A1E1WQL9 A0A2W1BYE8 I4DP89 D6WTI1 V5GPC0 A0A1L8D967 A0A1L8D949 N6TID4 A0A067RD41 A0A1Q3FBX3 U5EVY5 A0A1W4XRD5 A0A1B0CS20 A0A023EPS3 B0XHY4 Q1HQJ2 A0A182MN20 T1EAH5 A0A182VYE2 A0A182XWI1 A0A182LQQ1 A0A2J7QBS7 A0A2J7QBT5 T1DRV1 A0A2J7QBS0 A0A336MTE9 A0A182FQN7 A0A182WZI9 Q7QK67 A0A0K8U137 A0A0K8W6F8 A0A2M4AI20 A0A2M4BR45 A0A0K8TMF3 A0A0A1XN97 W5J4W1 A0A0A1WEZ0 A0A182TD98 A0A034WEU8 A0A034WGE6 A0A182HZN8 A0A1A9XDJ3 A0A1B0C4H0 A0A1A9ZGF1 W8BUF6 W8C161 A0A182PEB3 A0A182IM96 A0A2M3Z6T9 A0A2M3ZHY3 D3TMP1 Q8MTA2 Q8IMR3 A0A336MJU1 H8F4V3 Q6NNZ6 Q9VBC5 Q8IMR4 A0A1A9WSG8 A0A336MM83 A0A182Q771 A0A182NMW1 A0A0L0CQF8 A0A1B6E7H8 B4NKN4 A0A1Y1L7J5 A0A1Y1L7H1 A0A336MDY0 A0A0M4EJ72 A0A1I8Q7P3 T1JB28 A0A1I8Q7S5 A0A0Q9WTC3 A0A0P4WE48 B4LWB2 A0A0Q9WHG8 T1PCH3 A0A0P9CAS5 A0A1I8MJV6 A0A1B6IJP9 A0A0Q9X2V1 B3M3C8 A0A0Q9X2G0 B4K8Y8 A0A0P8YNK6 A0A1I8MJW2 A0A1W4VM29 A0A1W4VLC4 A0A1W4V8Y5 A0A3B0JL82 A0A3B0K004 B4JY65 A0A3B0K7W4 A0A0R1E554
A0A1E1WQL9 A0A2W1BYE8 I4DP89 D6WTI1 V5GPC0 A0A1L8D967 A0A1L8D949 N6TID4 A0A067RD41 A0A1Q3FBX3 U5EVY5 A0A1W4XRD5 A0A1B0CS20 A0A023EPS3 B0XHY4 Q1HQJ2 A0A182MN20 T1EAH5 A0A182VYE2 A0A182XWI1 A0A182LQQ1 A0A2J7QBS7 A0A2J7QBT5 T1DRV1 A0A2J7QBS0 A0A336MTE9 A0A182FQN7 A0A182WZI9 Q7QK67 A0A0K8U137 A0A0K8W6F8 A0A2M4AI20 A0A2M4BR45 A0A0K8TMF3 A0A0A1XN97 W5J4W1 A0A0A1WEZ0 A0A182TD98 A0A034WEU8 A0A034WGE6 A0A182HZN8 A0A1A9XDJ3 A0A1B0C4H0 A0A1A9ZGF1 W8BUF6 W8C161 A0A182PEB3 A0A182IM96 A0A2M3Z6T9 A0A2M3ZHY3 D3TMP1 Q8MTA2 Q8IMR3 A0A336MJU1 H8F4V3 Q6NNZ6 Q9VBC5 Q8IMR4 A0A1A9WSG8 A0A336MM83 A0A182Q771 A0A182NMW1 A0A0L0CQF8 A0A1B6E7H8 B4NKN4 A0A1Y1L7J5 A0A1Y1L7H1 A0A336MDY0 A0A0M4EJ72 A0A1I8Q7P3 T1JB28 A0A1I8Q7S5 A0A0Q9WTC3 A0A0P4WE48 B4LWB2 A0A0Q9WHG8 T1PCH3 A0A0P9CAS5 A0A1I8MJV6 A0A1B6IJP9 A0A0Q9X2V1 B3M3C8 A0A0Q9X2G0 B4K8Y8 A0A0P8YNK6 A0A1I8MJW2 A0A1W4VM29 A0A1W4VLC4 A0A1W4V8Y5 A0A3B0JL82 A0A3B0K004 B4JY65 A0A3B0K7W4 A0A0R1E554
Ontologies
GO
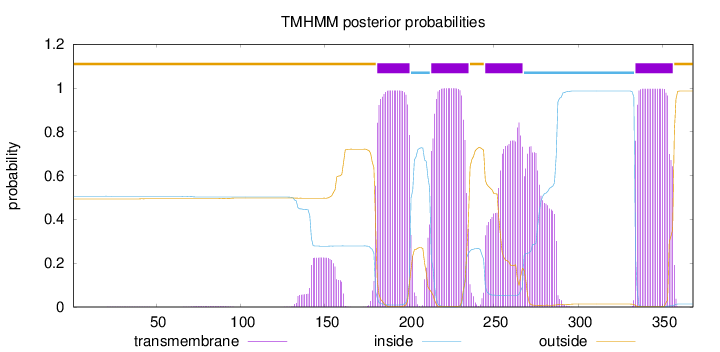
Topology
Length:
368
Number of predicted TMHs:
4
Exp number of AAs in TMHs:
94.48418
Exp number, first 60 AAs:
0.01908
Total prob of N-in:
0.50628
outside
1 - 180
TMhelix
181 - 200
inside
201 - 212
TMhelix
213 - 235
outside
236 - 244
TMhelix
245 - 267
inside
268 - 333
TMhelix
334 - 356
outside
357 - 368
Population Genetic Test Statistics
Pi
319.808884
Theta
212.545065
Tajima's D
1.639851
CLR
0.058184
CSRT
0.817909104544773
Interpretation
Uncertain
