Gene
KWMTBOMO14922
Pre Gene Modal
BGIBMGA000791
Annotation
PREDICTED:_serine/threonine-protein_kinase_D1_[Bombyx_mori]
Full name
Serine/threonine-protein kinase
+ More
Serine/threonine-protein kinase D1
Serine/threonine-protein kinase D1
Alternative Name
Protein kinase C mu type
Protein kinase D
nPKC-D1
nPKC-mu
Protein kinase D
nPKC-D1
nPKC-mu
Location in the cell
Nuclear Reliability : 2.777
Sequence
CDS
ATGTGCCCCAAACCGAGGTCGGGCAATCGAACGGTAGATGAGCACGTCAGCACGCGACGGGTGCCTGAATCGGAATGTTGCGGGTTGAACTACCACAAGCGTTGCGCGGTGAAGGTTCCGTCGAACTGCGCGTCGGCGTGCGCGGGCGGGGCGGCGGGCGGTGCGGGGGGCGCGGGCAGCACGCGGCGCCCCTCCGCCGCCGCCCCCCGCAGCCCGTCGCGCTCCTCCCAGCACAGCCACGCCTCGCACGACGACTCGCTGGTGGTAAACAAAAGAAGCAAATCCCCTTCTCGTGTGGTCTGGCCTCTGGGCCAAGGCAGCTCCCTCGGCATTCCTCATACCTTCGAAGTCCACACGTATACCCGGCCTACGGTTTGTCGCCATTGCAAGAAACTACTTCGAGGACTGTTCAAGCAAGGTCTTCAGTGTCGAGATTGTCATTACAATGCGCACAGGAAGTGTCTGCCCTTCGTGCCTAAGGATTGTGGAGGGGAGATACGAGACCAGCACGGAGAAGCGGACAGCGGGACCGGCAGTGAGCTATCGGAGACAGCGCCATTGCTGGACGAGTCTGACGAGGAAGCAACTCCTGATCACGCCGATCGAAATAGCGTGGATGAGGAGGTACAACTACGAAGACAAAGGGATGAGGATTCGGATGAACCGCAACGTGACACACTGCCGGAGCGTTCGGCGAGCCGACCCAGCAGTTCATCGTCATCACCCAGCGCGAACATCCCGCTGATGAGGATAGTGCAGTCGGTGAAGCACACCAAGAAACGGGAGGGACAATGGCTTAAGGAGGGCTGGCTTGTTCACTTTACTAATAAGGATAAGACGCTGAGACGTCACTACTGGCGCCTGGACAGCAAGTCGATAACCCTGTTCACATCGGACCAGGGCACCAAGTACTACAAGGAGATACCGCTCAATGAGATCCTGGCGATCGACACGGCCAGGCAGCCGCAGTCAGAGGTGATGCACTGCTTCGAGCTGCGCACGGCCAACGTGGACTATCTGGTGGGCGCGGAGGCAGGCCCGGGCGGCGCCGTGCCCTCCTCCGGCCTCGGCGCCTACCTCGCGCGCCCCTGGGAGACCGCTCTGCGTCACGCGCTCATGCCCGTGACGCACCACACGCGTCACGCACATCACGCGGTGGATGGCGGGTCGGGGAGTAGCGGGGCCGAGGGCGGGGCTGAAGGCGGGGCTGCGGGGGCCGACGGCGGGGAGGGGGGCGCGGCCTGCGCGGAGGAGAGGGTGACGGATATGGCGCAGCTCTACCACATCCACCCCGACGAGGTGCTGGGGTCTGGCCAGTTTGGAATCGTATACGGCGGCCTGCACCGGAGGACGGACAGACCTGTCGCCATTAAAGTAATAGATAAACTCCGCTTCCCGACCAAACAAGAGGCGCAACTGAAGAATGAAGTGGCCATTCTTCAGAACCTGTCCCACCCCGGCGTCGTGAACCTCGAGAGGATGTTCGAGACCCCGGAGCGGATCTTCGTGGTGATGGAGAAGCTGAAGGGGGACATGCTGGAGATGATCCTCTCGCACGAGAGGGGAAGACTCACCGAGCGAATCACCAAGTTCCTGGTCGCACAGATCCTGGTGGCCCTGAAGCATCTCCACGAGAAGAACATCGTGCACTGCGACCTGAAGCCGGAGAACGTGCTGCTGTCCACGGACGACGAGTTCCCGCAAGTCAAGCTCTGCGACTTCGGGTTCGCGCGCATCATCGGCGAGAAGTCATTCCGGAGATCCGTCGTGGGGACGCCGGCCTACCTGGCTCCGGAAGTGTTGCGCAACAAGGGCTACAACCGCTCGCTCGACATGTGGTCCGTGGGCGTCATCATCTACGTGTCGCTGTCCGGGACGTTCCCCTTCAACGAGGACGAGGACATCAACGAGCAGATACAGAACGCTGCCTTCATGTACCCGCCCGCGCCCTGGAGGGAGATCTCCGCCGAGGCAATCGACCTCATAAACAACCTGTTGCAAGTGAAGCAACGCAAGCGGCTGTCCGTGGACAAGTCCGTGTCCCACTGCTGGCTGCAGACGCGCGAGGCATGGCAGGACCTGCGCGCCCTCGAGCGCCGAGTCGGCACCCGCTACCTCACGCACCCCAGCGACGACGCACGCTGGACACGCTGA
Protein
MCPKPRSGNRTVDEHVSTRRVPESECCGLNYHKRCAVKVPSNCASACAGGAAGGAGGAGSTRRPSAAAPRSPSRSSQHSHASHDDSLVVNKRSKSPSRVVWPLGQGSSLGIPHTFEVHTYTRPTVCRHCKKLLRGLFKQGLQCRDCHYNAHRKCLPFVPKDCGGEIRDQHGEADSGTGSELSETAPLLDESDEEATPDHADRNSVDEEVQLRRQRDEDSDEPQRDTLPERSASRPSSSSSSPSANIPLMRIVQSVKHTKKREGQWLKEGWLVHFTNKDKTLRRHYWRLDSKSITLFTSDQGTKYYKEIPLNEILAIDTARQPQSEVMHCFELRTANVDYLVGAEAGPGGAVPSSGLGAYLARPWETALRHALMPVTHHTRHAHHAVDGGSGSSGAEGGAEGGAAGADGGEGGAACAEERVTDMAQLYHIHPDEVLGSGQFGIVYGGLHRRTDRPVAIKVIDKLRFPTKQEAQLKNEVAILQNLSHPGVVNLERMFETPERIFVVMEKLKGDMLEMILSHERGRLTERITKFLVAQILVALKHLHEKNIVHCDLKPENVLLSTDDEFPQVKLCDFGFARIIGEKSFRRSVVGTPAYLAPEVLRNKGYNRSLDMWSVGVIIYVSLSGTFPFNEDEDINEQIQNAAFMYPPAPWREISAEAIDLINNLLQVKQRKRLSVDKSVSHCWLQTREAWQDLRALERRVGTRYLTHPSDDARWTR
Summary
Description
Serine/threonine-protein kinase that converts transient diacylglycerol (DAG) signals into prolonged physiological effects downstream of PKC, and is involved in the regulation of MAPK8/JNK1 and Ras signaling, Golgi membrane integrity and trafficking, cell survival through NF-kappa-B activation, cell migration, cell differentiation by mediating HDAC7 nuclear export, cell proliferation via MAPK1/3 (ERK1/2) signaling, and plays a role in cardiac hypertrophy, VEGFA-induced angiogenesis, genotoxic-induced apoptosis and flagellin-stimulated inflammatory response. Phosphorylates the epidermal growth factor receptor (EGFR) on dual threonine residues, which leads to the suppression of epidermal growth factor (EGF)-induced MAPK8/JNK1 activation and subsequent JUN phosphorylation. Phosphorylates RIN1, inducing RIN1 binding to 14-3-3 proteins YWHAB, YWHAE and YWHAZ and increased competition with RAF1 for binding to GTP-bound form of Ras proteins (NRAS, HRAS and KRAS). Acts downstream of the heterotrimeric G-protein beta/gamma-subunit complex to maintain the structural integrity of the Golgi membranes, and is required for protein transport along the secretory pathway. In the trans-Golgi network (TGN), regulates the fission of transport vesicles that are on their way to the plasma membrane. May act by activating the lipid kinase phosphatidylinositol 4-kinase beta (PI4KB) at the TGN for the local synthesis of phosphorylated inositol lipids, which induces a sequential production of DAG, phosphatidic acid (PA) and lyso-PA (LPA) that are necessary for membrane fission and generation of specific transport carriers to the cell surface. Under oxidative stress, is phosphorylated at Tyr-469 via SRC-ABL1 and contributes to cell survival by activating IKK complex and subsequent nuclear translocation and activation of NFKB1. Involved in cell migration by regulating integrin alpha-5/beta-3 recycling and promoting its recruitment in newly forming focal adhesion. In osteoblast differentiation, mediates the bone morphogenetic protein 2 (BMP2)-induced nuclear export of HDAC7, which results in the inhibition of HDAC7 transcriptional repression of RUNX2. In neurons, plays an important role in neuronal polarity by regulating the biogenesis of TGN-derived dendritic vesicles, and is involved in the maintenance of dendritic arborization and Golgi structure in hippocampal cells. May potentiate mitogenesis induced by the neuropeptide bombesin or vasopressin by mediating an increase in the duration of MAPK1/3 (ERK1/2) signaling, which leads to accumulation of immediate-early gene products including FOS that stimulate cell cycle progression. Plays an important role in the proliferative response induced by low calcium in keratinocytes, through sustained activation of MAPK1/3 (ERK1/2) pathway. Downstream of novel PKC signaling, plays a role in cardiac hypertrophy by phosphorylating HDAC5, which in turn triggers XPO1/CRM1-dependent nuclear export of HDAC5, MEF2A transcriptional activation and induction of downstream target genes that promote myocyte hypertrophy and pathological cardiac remodeling. Mediates cardiac troponin I (TNNI3) phosphorylation at the PKA sites, which results in reduced myofilament calcium sensitivity, and accelerated crossbridge cycling kinetics. The PRKD1-HDAC5 pathway is also involved in angiogenesis by mediating VEGFA-induced specific subset of gene expression, cell migration, and tube formation. In response to VEGFA, is necessary and required for HDAC7 phosphorylation which induces HDAC7 nuclear export and endothelial cell proliferation and migration. During apoptosis induced by cytarabine and other genotoxic agents, PRKD1 is cleaved by caspase-3 at Asp-378, resulting in activation of its kinase function and increased sensitivity of cells to the cytotoxic effects of genotoxic agents. In epithelial cells, is required for transducing flagellin-stimulated inflammatory responses by binding and phosphorylating TLR5, which contributes to MAPK14/p38 activation and production of inflammatory cytokines. May play a role in inflammatory response by mediating activation of NF-kappa-B. May be involved in pain transmission by directly modulating TRPV1 receptor. Plays a role in activated KRAS-mediated stabilization of ZNF304 in colorectal cancer (CRC) cells (By similarity). Regulates nuclear translocation of transcription factor TFEB in macrophages upon live S.enterica infection (By similarity).
Catalytic Activity
ATP + L-threonyl-[protein] = ADP + H(+) + O-phospho-L-threonyl-[protein]
ATP + L-seryl-[protein] = ADP + H(+) + O-phospho-L-seryl-[protein]
ATP + L-seryl-[protein] = ADP + H(+) + O-phospho-L-seryl-[protein]
Cofactor
Mg(2+)
Subunit
Interacts (via N-terminus) with ADAP1/CENTA1. Interacts with MAPK13. Interacts with DAPK1 in an oxidative stress-regulated manner. Interacts with USP28; the interaction induces phosphorylation of USP28 and activated KRAS-mediated stabilization of ZNF304 (By similarity). Interacts with AKAP13 (via C-terminal domain) (By similarity).
Similarity
Belongs to the protein kinase superfamily. CAMK Ser/Thr protein kinase family.
Belongs to the protein kinase superfamily. CAMK Ser/Thr protein kinase family. PKD subfamily.
Belongs to the protein kinase superfamily. CAMK Ser/Thr protein kinase family. PKD subfamily.
Keywords
Angiogenesis
Apoptosis
ATP-binding
Cell membrane
Complete proteome
Cytoplasm
Differentiation
Golgi apparatus
Immunity
Inflammatory response
Innate immunity
Kinase
Magnesium
Membrane
Metal-binding
Neurogenesis
Nucleotide-binding
Phosphoprotein
Reference proteome
Repeat
Transferase
Zinc
Zinc-finger
Feature
chain Serine/threonine-protein kinase
Uniprot
H9IU61
A0A3S2M2I1
A0A194PM08
A0A2H1W480
A0A026VV67
Q17GS6
+ More
A0A1S4F342 A0A2W1BP91 U5Q3U4 A0A139WES9 A0A182H9Q1 N6SYX1 F4X1E6 A0A195FRL9 A0A2M4ACC2 A0A2M4ACC3 A0A151WX40 A0A2M4ADM9 A0A195BK94 A0A232EIM1 A0A2M3ZGT9 A0A182M5B1 A0A195DLS6 A0A195CVA2 A0A158NA49 A0A182FJE3 A0A2M4BCQ5 A0A182KA76 A0A1Q3FXD5 A0A182YCM5 E2BAK2 A0A182RZ35 W5JDN3 A0A139WF71 A0A2M4CPE6 A0A182VVP2 A0A088A8E8 A0A0C9QSD0 A0A1W4WN85 A0A2A3E987 A0A1B6DCZ5 A0A0N0BHP1 A0A310SWG2 A0A0L7QR25 A0A182N023 A0A1L8DKS6 A0A154PFX7 A0A1L8DL51 A0A0A9XVX8 A0A1Y1M919 A0A2J7Q1W3 A0A224XJG3 A0A023F3P7 A0A0P4VZQ6 A0A3R7SUA9 A0A0N7ZCN2 A0A2I0UTW7 A0A226ETX1 A0A1V4KFB8 G1TPY2 B4M405 E9G2E3 B4K940 B4JF29 A0A341C9W6 A0A2Y9LQ04 W5MU06 A0A1U7QP75 W5MTZ8 A0A2Y9THQ4 A0A2K5KM13 A0A384A039 A0A0P5C5F1 A0A2Y9TFV5 A0A0P5GEL0 A0A0P6HXR5 A0A0N8AU91 A0A2K6K1X8 A0A2Y9DKX1 A0A2K5KM23 F6R5K3 A0A0P6DSL2 U6DFF8 A0A340YCC6 F7I1N3 A0A0N8ATC7 A0A0P6FU07 W5P4Q9 A0A0P6BHC7 A0A0P5NFS2 A0A2K5EB98 A0A2I3SNQ8 A0A2K6V3G4 A0A0P5W1R9 A0A2K5WZ23 A0A2K5WZC4 A0A2K5EBA4 A0A1D5QQ16 A0A0P5A541 Q9WTQ1
A0A1S4F342 A0A2W1BP91 U5Q3U4 A0A139WES9 A0A182H9Q1 N6SYX1 F4X1E6 A0A195FRL9 A0A2M4ACC2 A0A2M4ACC3 A0A151WX40 A0A2M4ADM9 A0A195BK94 A0A232EIM1 A0A2M3ZGT9 A0A182M5B1 A0A195DLS6 A0A195CVA2 A0A158NA49 A0A182FJE3 A0A2M4BCQ5 A0A182KA76 A0A1Q3FXD5 A0A182YCM5 E2BAK2 A0A182RZ35 W5JDN3 A0A139WF71 A0A2M4CPE6 A0A182VVP2 A0A088A8E8 A0A0C9QSD0 A0A1W4WN85 A0A2A3E987 A0A1B6DCZ5 A0A0N0BHP1 A0A310SWG2 A0A0L7QR25 A0A182N023 A0A1L8DKS6 A0A154PFX7 A0A1L8DL51 A0A0A9XVX8 A0A1Y1M919 A0A2J7Q1W3 A0A224XJG3 A0A023F3P7 A0A0P4VZQ6 A0A3R7SUA9 A0A0N7ZCN2 A0A2I0UTW7 A0A226ETX1 A0A1V4KFB8 G1TPY2 B4M405 E9G2E3 B4K940 B4JF29 A0A341C9W6 A0A2Y9LQ04 W5MU06 A0A1U7QP75 W5MTZ8 A0A2Y9THQ4 A0A2K5KM13 A0A384A039 A0A0P5C5F1 A0A2Y9TFV5 A0A0P5GEL0 A0A0P6HXR5 A0A0N8AU91 A0A2K6K1X8 A0A2Y9DKX1 A0A2K5KM23 F6R5K3 A0A0P6DSL2 U6DFF8 A0A340YCC6 F7I1N3 A0A0N8ATC7 A0A0P6FU07 W5P4Q9 A0A0P6BHC7 A0A0P5NFS2 A0A2K5EB98 A0A2I3SNQ8 A0A2K6V3G4 A0A0P5W1R9 A0A2K5WZ23 A0A2K5WZC4 A0A2K5EBA4 A0A1D5QQ16 A0A0P5A541 Q9WTQ1
EC Number
2.7.11.13
Pubmed
19121390
26354079
24508170
30249741
17510324
28756777
+ More
24146884 18362917 19820115 26483478 23537049 21719571 28648823 21347285 25244985 20798317 20920257 23761445 25401762 26823975 28004739 25474469 27129103 21993624 17994087 21292972 18464734 20809919 16136131 17431167 15057822 10412981 15514163 15145937 15367659 18784310 22673903
24146884 18362917 19820115 26483478 23537049 21719571 28648823 21347285 25244985 20798317 20920257 23761445 25401762 26823975 28004739 25474469 27129103 21993624 17994087 21292972 18464734 20809919 16136131 17431167 15057822 10412981 15514163 15145937 15367659 18784310 22673903
EMBL
BABH01034181
BABH01034182
RSAL01000057
RVE49778.1
KQ459604
KPI92140.1
+ More
ODYU01006241 SOQ47900.1 KK107796 QOIP01000013 EZA47622.1 RLU15226.1 CH477256 EAT45861.1 KZ149945 PZC76788.1 KC896834 AGY49605.1 KQ971354 KYB26446.1 JXUM01121273 KQ566481 KXJ70098.1 APGK01051641 KB741198 ENN72969.1 GL888529 EGI59741.1 KQ981305 KYN42932.1 GGFK01005123 MBW38444.1 GGFK01005124 MBW38445.1 KQ982686 KYQ52327.1 GGFK01005562 MBW38883.1 KQ976455 KYM85112.1 NNAY01004210 OXU18195.1 GGFM01006944 MBW27695.1 AXCM01006825 KQ980734 KYN13796.1 KQ977276 KYN04467.1 ADTU01009967 GGFJ01001662 MBW50803.1 GFDL01002933 JAV32112.1 GL446764 EFN87279.1 ADMH02001467 ETN62472.1 KYB26447.1 GGFL01002883 MBW67061.1 GBYB01006644 JAG76411.1 KZ288325 PBC28034.1 GEDC01013749 JAS23549.1 KQ435747 KOX76512.1 KQ759883 OAD62173.1 KQ414786 KOC60946.1 GFDF01007015 JAV07069.1 KQ434896 KZC10786.1 GFDF01007014 JAV07070.1 GBHO01020581 GDHC01016161 JAG23023.1 JAQ02468.1 GEZM01036985 GEZM01036984 JAV82292.1 NEVH01019377 PNF22567.1 GFTR01007694 JAW08732.1 GBBI01002692 JAC16020.1 GDKW01001588 JAI55007.1 QCYY01001774 ROT75419.1 GDRN01063911 JAI64920.1 KZ505637 PKU49503.1 LNIX01000002 OXA60504.1 LSYS01003385 OPJ83132.1 AAGW02023970 AAGW02023971 AAGW02023972 AAGW02023973 AAGW02023974 AAGW02023975 CH940652 EDW59366.2 GL732530 EFX86325.1 CH933806 EDW14453.2 CH916369 EDV93310.1 AHAT01003849 GDIP01179515 JAJ43887.1 GDIQ01243592 JAK08133.1 GDIQ01020520 JAN74217.1 GDIQ01242331 JAK09394.1 AAPN01002929 AAPN01002930 GDIQ01087885 JAN06852.1 HAAF01009517 CCP81341.1 GDIQ01244843 JAK06882.1 GDIQ01053805 JAN40932.1 AMGL01042243 AMGL01042244 AMGL01042245 AMGL01042246 AMGL01042247 AMGL01042248 AMGL01042249 AMGL01042250 GDIP01014565 JAM89150.1 GDIQ01149470 JAL02256.1 AACZ04059434 AACZ04059435 AACZ04059436 AACZ04059437 AACZ04059438 GDIP01093084 JAM10631.1 AQIA01064312 AQIA01064313 AQIA01064314 AQIA01064315 AQIA01064316 AQIA01064317 AQIA01064318 AQIA01064319 AQIA01064320 AQIA01064321 JSUE03039390 JSUE03039391 JSUE03039392 JSUE03039393 JSUE03039394 JSUE03039395 JSUE03039396 JSUE03039397 GDIP01205016 JAJ18386.1 AABR03048549 AABR03048623 AABR03048825 AABR03049215 AABR03049278 AABR03050321 AABR03052344 AB020616
ODYU01006241 SOQ47900.1 KK107796 QOIP01000013 EZA47622.1 RLU15226.1 CH477256 EAT45861.1 KZ149945 PZC76788.1 KC896834 AGY49605.1 KQ971354 KYB26446.1 JXUM01121273 KQ566481 KXJ70098.1 APGK01051641 KB741198 ENN72969.1 GL888529 EGI59741.1 KQ981305 KYN42932.1 GGFK01005123 MBW38444.1 GGFK01005124 MBW38445.1 KQ982686 KYQ52327.1 GGFK01005562 MBW38883.1 KQ976455 KYM85112.1 NNAY01004210 OXU18195.1 GGFM01006944 MBW27695.1 AXCM01006825 KQ980734 KYN13796.1 KQ977276 KYN04467.1 ADTU01009967 GGFJ01001662 MBW50803.1 GFDL01002933 JAV32112.1 GL446764 EFN87279.1 ADMH02001467 ETN62472.1 KYB26447.1 GGFL01002883 MBW67061.1 GBYB01006644 JAG76411.1 KZ288325 PBC28034.1 GEDC01013749 JAS23549.1 KQ435747 KOX76512.1 KQ759883 OAD62173.1 KQ414786 KOC60946.1 GFDF01007015 JAV07069.1 KQ434896 KZC10786.1 GFDF01007014 JAV07070.1 GBHO01020581 GDHC01016161 JAG23023.1 JAQ02468.1 GEZM01036985 GEZM01036984 JAV82292.1 NEVH01019377 PNF22567.1 GFTR01007694 JAW08732.1 GBBI01002692 JAC16020.1 GDKW01001588 JAI55007.1 QCYY01001774 ROT75419.1 GDRN01063911 JAI64920.1 KZ505637 PKU49503.1 LNIX01000002 OXA60504.1 LSYS01003385 OPJ83132.1 AAGW02023970 AAGW02023971 AAGW02023972 AAGW02023973 AAGW02023974 AAGW02023975 CH940652 EDW59366.2 GL732530 EFX86325.1 CH933806 EDW14453.2 CH916369 EDV93310.1 AHAT01003849 GDIP01179515 JAJ43887.1 GDIQ01243592 JAK08133.1 GDIQ01020520 JAN74217.1 GDIQ01242331 JAK09394.1 AAPN01002929 AAPN01002930 GDIQ01087885 JAN06852.1 HAAF01009517 CCP81341.1 GDIQ01244843 JAK06882.1 GDIQ01053805 JAN40932.1 AMGL01042243 AMGL01042244 AMGL01042245 AMGL01042246 AMGL01042247 AMGL01042248 AMGL01042249 AMGL01042250 GDIP01014565 JAM89150.1 GDIQ01149470 JAL02256.1 AACZ04059434 AACZ04059435 AACZ04059436 AACZ04059437 AACZ04059438 GDIP01093084 JAM10631.1 AQIA01064312 AQIA01064313 AQIA01064314 AQIA01064315 AQIA01064316 AQIA01064317 AQIA01064318 AQIA01064319 AQIA01064320 AQIA01064321 JSUE03039390 JSUE03039391 JSUE03039392 JSUE03039393 JSUE03039394 JSUE03039395 JSUE03039396 JSUE03039397 GDIP01205016 JAJ18386.1 AABR03048549 AABR03048623 AABR03048825 AABR03049215 AABR03049278 AABR03050321 AABR03052344 AB020616
Proteomes
UP000005204
UP000283053
UP000053268
UP000053097
UP000279307
UP000008820
+ More
UP000007266 UP000069940 UP000249989 UP000019118 UP000007755 UP000078541 UP000075809 UP000078540 UP000215335 UP000075883 UP000078492 UP000078542 UP000005205 UP000069272 UP000075881 UP000076408 UP000008237 UP000075900 UP000000673 UP000075920 UP000005203 UP000192223 UP000242457 UP000053105 UP000053825 UP000075884 UP000076502 UP000235965 UP000283509 UP000198287 UP000190648 UP000001811 UP000008792 UP000000305 UP000009192 UP000001070 UP000252040 UP000248483 UP000018468 UP000189706 UP000233060 UP000261681 UP000248484 UP000233180 UP000248480 UP000002279 UP000265300 UP000008225 UP000002356 UP000233020 UP000002277 UP000233220 UP000233100 UP000006718 UP000002494
UP000007266 UP000069940 UP000249989 UP000019118 UP000007755 UP000078541 UP000075809 UP000078540 UP000215335 UP000075883 UP000078492 UP000078542 UP000005205 UP000069272 UP000075881 UP000076408 UP000008237 UP000075900 UP000000673 UP000075920 UP000005203 UP000192223 UP000242457 UP000053105 UP000053825 UP000075884 UP000076502 UP000235965 UP000283509 UP000198287 UP000190648 UP000001811 UP000008792 UP000000305 UP000009192 UP000001070 UP000252040 UP000248483 UP000018468 UP000189706 UP000233060 UP000261681 UP000248484 UP000233180 UP000248480 UP000002279 UP000265300 UP000008225 UP000002356 UP000233020 UP000002277 UP000233220 UP000233100 UP000006718 UP000002494
Interpro
SUPFAM
SSF56112
SSF56112
Gene 3D
CDD
ProteinModelPortal
H9IU61
A0A3S2M2I1
A0A194PM08
A0A2H1W480
A0A026VV67
Q17GS6
+ More
A0A1S4F342 A0A2W1BP91 U5Q3U4 A0A139WES9 A0A182H9Q1 N6SYX1 F4X1E6 A0A195FRL9 A0A2M4ACC2 A0A2M4ACC3 A0A151WX40 A0A2M4ADM9 A0A195BK94 A0A232EIM1 A0A2M3ZGT9 A0A182M5B1 A0A195DLS6 A0A195CVA2 A0A158NA49 A0A182FJE3 A0A2M4BCQ5 A0A182KA76 A0A1Q3FXD5 A0A182YCM5 E2BAK2 A0A182RZ35 W5JDN3 A0A139WF71 A0A2M4CPE6 A0A182VVP2 A0A088A8E8 A0A0C9QSD0 A0A1W4WN85 A0A2A3E987 A0A1B6DCZ5 A0A0N0BHP1 A0A310SWG2 A0A0L7QR25 A0A182N023 A0A1L8DKS6 A0A154PFX7 A0A1L8DL51 A0A0A9XVX8 A0A1Y1M919 A0A2J7Q1W3 A0A224XJG3 A0A023F3P7 A0A0P4VZQ6 A0A3R7SUA9 A0A0N7ZCN2 A0A2I0UTW7 A0A226ETX1 A0A1V4KFB8 G1TPY2 B4M405 E9G2E3 B4K940 B4JF29 A0A341C9W6 A0A2Y9LQ04 W5MU06 A0A1U7QP75 W5MTZ8 A0A2Y9THQ4 A0A2K5KM13 A0A384A039 A0A0P5C5F1 A0A2Y9TFV5 A0A0P5GEL0 A0A0P6HXR5 A0A0N8AU91 A0A2K6K1X8 A0A2Y9DKX1 A0A2K5KM23 F6R5K3 A0A0P6DSL2 U6DFF8 A0A340YCC6 F7I1N3 A0A0N8ATC7 A0A0P6FU07 W5P4Q9 A0A0P6BHC7 A0A0P5NFS2 A0A2K5EB98 A0A2I3SNQ8 A0A2K6V3G4 A0A0P5W1R9 A0A2K5WZ23 A0A2K5WZC4 A0A2K5EBA4 A0A1D5QQ16 A0A0P5A541 Q9WTQ1
A0A1S4F342 A0A2W1BP91 U5Q3U4 A0A139WES9 A0A182H9Q1 N6SYX1 F4X1E6 A0A195FRL9 A0A2M4ACC2 A0A2M4ACC3 A0A151WX40 A0A2M4ADM9 A0A195BK94 A0A232EIM1 A0A2M3ZGT9 A0A182M5B1 A0A195DLS6 A0A195CVA2 A0A158NA49 A0A182FJE3 A0A2M4BCQ5 A0A182KA76 A0A1Q3FXD5 A0A182YCM5 E2BAK2 A0A182RZ35 W5JDN3 A0A139WF71 A0A2M4CPE6 A0A182VVP2 A0A088A8E8 A0A0C9QSD0 A0A1W4WN85 A0A2A3E987 A0A1B6DCZ5 A0A0N0BHP1 A0A310SWG2 A0A0L7QR25 A0A182N023 A0A1L8DKS6 A0A154PFX7 A0A1L8DL51 A0A0A9XVX8 A0A1Y1M919 A0A2J7Q1W3 A0A224XJG3 A0A023F3P7 A0A0P4VZQ6 A0A3R7SUA9 A0A0N7ZCN2 A0A2I0UTW7 A0A226ETX1 A0A1V4KFB8 G1TPY2 B4M405 E9G2E3 B4K940 B4JF29 A0A341C9W6 A0A2Y9LQ04 W5MU06 A0A1U7QP75 W5MTZ8 A0A2Y9THQ4 A0A2K5KM13 A0A384A039 A0A0P5C5F1 A0A2Y9TFV5 A0A0P5GEL0 A0A0P6HXR5 A0A0N8AU91 A0A2K6K1X8 A0A2Y9DKX1 A0A2K5KM23 F6R5K3 A0A0P6DSL2 U6DFF8 A0A340YCC6 F7I1N3 A0A0N8ATC7 A0A0P6FU07 W5P4Q9 A0A0P6BHC7 A0A0P5NFS2 A0A2K5EB98 A0A2I3SNQ8 A0A2K6V3G4 A0A0P5W1R9 A0A2K5WZ23 A0A2K5WZC4 A0A2K5EBA4 A0A1D5QQ16 A0A0P5A541 Q9WTQ1
PDB
2YCR
E-value=1.16055e-48,
Score=490
Ontologies
GO
GO:0035556
GO:0046872
GO:0004697
GO:0005524
GO:0004674
GO:0005737
GO:0089700
GO:0016021
GO:0010837
GO:0005802
GO:0005911
GO:0005886
GO:0045669
GO:0005634
GO:0042307
GO:0005829
GO:0005938
GO:0050829
GO:0051279
GO:0048193
GO:1901490
GO:0045765
GO:0051092
GO:0006954
GO:0043123
GO:0007030
GO:0010595
GO:0001525
GO:0035924
GO:0046777
GO:0045087
GO:0010976
GO:0045766
GO:0006915
GO:0007265
GO:0031647
GO:0060548
GO:0034599
GO:0004672
GO:0006468
GO:0007154
GO:0016020
GO:0005112
GO:0015930
PANTHER
Topology
Subcellular location
Cytoplasm
Cell membrane Translocation to the cell membrane is required for kinase activation. With evidence from 2 publications.
Golgi apparatus Translocation to the cell membrane is required for kinase activation. With evidence from 2 publications.
trans-Golgi network Translocation to the cell membrane is required for kinase activation. With evidence from 2 publications.
Cell membrane Translocation to the cell membrane is required for kinase activation. With evidence from 2 publications.
Golgi apparatus Translocation to the cell membrane is required for kinase activation. With evidence from 2 publications.
trans-Golgi network Translocation to the cell membrane is required for kinase activation. With evidence from 2 publications.
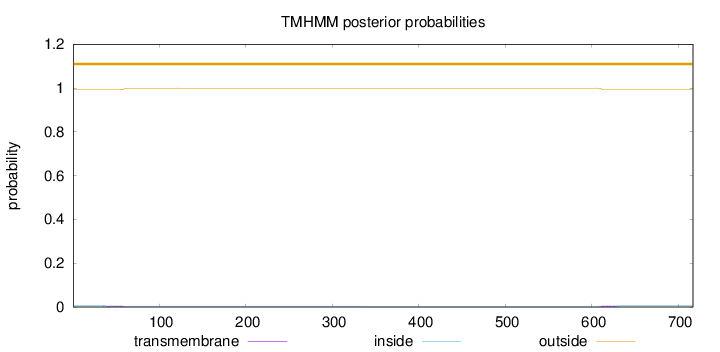
Length:
717
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.237960000000001
Exp number, first 60 AAs:
0.11891
Total prob of N-in:
0.00643
outside
1 - 717
Population Genetic Test Statistics
Pi
243.921366
Theta
172.716745
Tajima's D
1.19921
CLR
0.484629
CSRT
0.712764361781911
Interpretation
Uncertain
