Gene
KWMTBOMO14920
Pre Gene Modal
BGIBMGA000790
Annotation
PREDICTED:_serine/threonine-protein_kinase_D1_[Bombyx_mori]
Full name
Serine/threonine-protein kinase
Location in the cell
Extracellular Reliability : 2.452
Sequence
CDS
ATGGCGGCTGGAGACGCGATGGAGGACGAGGTGACGTTCATGTTCCAGCTGGGAGTGCTCCGGGACGCGGTCTCGGCGCCAGCGCATCTCCTCACGCTGGCACACCTCAAGGAGCTAGCAGTCAAATTCGTCCACGAAAAGATCCCGGACAATGGTCTGAATCGTCTAGCGGATCGGATCCTGTTGTTCAGACATGACTACTGTTCGCCCAACGTTCTCCAACTCCTCAACTCCGGGACAGACGTCGTGGACGAGACCCTCGTGGAGATCGTGCTCACAGCTAACCCGCTAATATGCGACGTCGGCACGGAGAGTTTGCCACTGATGCGGCCCCACACGCTCGCCGTGCACTCGTACAAGGCGCCGACCTTCTGCGACTTCTGCGGGGAGATGCTCTTCGGTCTCGTGCGACAAGGACTCAAGTGTGAAGTACTATTTGTTGAAATATTAATAAAAATGTCTAAAAATGTCATGTCATATCGAAAAGAAGAAAAGAAGAAATGTTAA
Protein
MAAGDAMEDEVTFMFQLGVLRDAVSAPAHLLTLAHLKELAVKFVHEKIPDNGLNRLADRILLFRHDYCSPNVLQLLNSGTDVVDETLVEIVLTANPLICDVGTESLPLMRPHTLAVHSYKAPTFCDFCGEMLFGLVRQGLKCEVLFVEILIKMSKNVMSYRKEEKKKC
Summary
Catalytic Activity
ATP + L-threonyl-[protein] = ADP + H(+) + O-phospho-L-threonyl-[protein]
Cofactor
Mg(2+)
Similarity
Belongs to the protein kinase superfamily. CAMK Ser/Thr protein kinase family.
Uniprot
H9IU60
A0A2H1X0J4
A0A2W1BP91
A0A194RHM1
A0A194PM08
A0A212EHA8
+ More
S4P1R1 A0A1Q3F8R2 A0A1Q3F9U0 B0WHS9 Q17GS6 A0A1S4F342 A0A1Q3FXD5 A0A182H9Q1 A0A1B6JF00 A0A0B4KGF0 A0A1B6GPA7 A0A336M6P4 A0A1B6HYA8 A0A0L0C6W2 A0A0K8UC92 W8AX18 D6WQI6 A0A182V8R4 A0A139WEP4 A0A0A1WE52 A0A1I8NM65 A0A034VCP3 T1PPM3 A0A182SIT0 A0A1B0CGD3 B4N9C8 A0A1Y1MCH3 B3NZ72 B4K940 B4JF29 A0A1I8MGR5 B4G5C4 A0A1W4U8S3 A0A1L8DKS6 A0A1L8DL51 Q9VE91 A0A0B4KHC3 B4QUG0 A0A3B0KS02 B4M405 B4IB64 A0A1Y1M9B7 B4PLA9 B5DY27 B3LWQ0 A0A2M4ACC3 A0A182PTP1 A0A182M5B1 A0A139WES9 A0A1Y1M919 A0A182N023 U5Q3U4 A0A340TB55 Q7PS40 W5JDN3 A0A182HIP5 A0A182YCM5 A0A2M4BCQ5 A0A182FJE3 A0A1Y1M9N6 A0A158NA49 F4X1E6 A0A195BK94 A0A1A9VEP9 A0A182KA76 A0A1B0F9Q4 A0A195FRL9 U4UQ62 A0A0A9XVX8 A0A084VJ01 A0A182QBL7 A0A182J288 A0A2J7Q1W3 E2BAK2 A0A2R7WVP8 A0A2A3E987 A0A0J7NWI4 A0A0N0BHP1 A0A310SWG2 A0A088A8E8 A0A1B0BJC4 A0A151WX40 N6SYX1 A0A195DLS6 A0A195CVA2 A0A154PFX7 A0A026VV67 A0A0L7QR25 A0A1A9XZB5 A0A2P8YZX4 A0A1W4WN85 A0A1A9W3J6 A0A023F3P7 A0A232EIM1
S4P1R1 A0A1Q3F8R2 A0A1Q3F9U0 B0WHS9 Q17GS6 A0A1S4F342 A0A1Q3FXD5 A0A182H9Q1 A0A1B6JF00 A0A0B4KGF0 A0A1B6GPA7 A0A336M6P4 A0A1B6HYA8 A0A0L0C6W2 A0A0K8UC92 W8AX18 D6WQI6 A0A182V8R4 A0A139WEP4 A0A0A1WE52 A0A1I8NM65 A0A034VCP3 T1PPM3 A0A182SIT0 A0A1B0CGD3 B4N9C8 A0A1Y1MCH3 B3NZ72 B4K940 B4JF29 A0A1I8MGR5 B4G5C4 A0A1W4U8S3 A0A1L8DKS6 A0A1L8DL51 Q9VE91 A0A0B4KHC3 B4QUG0 A0A3B0KS02 B4M405 B4IB64 A0A1Y1M9B7 B4PLA9 B5DY27 B3LWQ0 A0A2M4ACC3 A0A182PTP1 A0A182M5B1 A0A139WES9 A0A1Y1M919 A0A182N023 U5Q3U4 A0A340TB55 Q7PS40 W5JDN3 A0A182HIP5 A0A182YCM5 A0A2M4BCQ5 A0A182FJE3 A0A1Y1M9N6 A0A158NA49 F4X1E6 A0A195BK94 A0A1A9VEP9 A0A182KA76 A0A1B0F9Q4 A0A195FRL9 U4UQ62 A0A0A9XVX8 A0A084VJ01 A0A182QBL7 A0A182J288 A0A2J7Q1W3 E2BAK2 A0A2R7WVP8 A0A2A3E987 A0A0J7NWI4 A0A0N0BHP1 A0A310SWG2 A0A088A8E8 A0A1B0BJC4 A0A151WX40 N6SYX1 A0A195DLS6 A0A195CVA2 A0A154PFX7 A0A026VV67 A0A0L7QR25 A0A1A9XZB5 A0A2P8YZX4 A0A1W4WN85 A0A1A9W3J6 A0A023F3P7 A0A232EIM1
EC Number
2.7.11.13
Pubmed
19121390
28756777
26354079
22118469
23622113
17510324
+ More
26483478 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 26108605 24495485 18362917 19820115 25830018 25348373 17994087 28004739 25315136 17550304 15632085 24146884 12364791 14747013 17210077 20920257 23761445 25244985 21347285 21719571 23537049 25401762 26823975 24438588 20798317 24508170 30249741 29403074 25474469 28648823
26483478 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 26108605 24495485 18362917 19820115 25830018 25348373 17994087 28004739 25315136 17550304 15632085 24146884 12364791 14747013 17210077 20920257 23761445 25244985 21347285 21719571 23537049 25401762 26823975 24438588 20798317 24508170 30249741 29403074 25474469 28648823
EMBL
BABH01034178
ODYU01012455
SOQ58772.1
KZ149945
PZC76788.1
KQ460205
+ More
KPJ17062.1 KQ459604 KPI92140.1 AGBW02014918 OWR40862.1 GAIX01007184 JAA85376.1 GFDL01011126 JAV23919.1 GFDL01010729 JAV24316.1 DS231939 EDS27946.1 CH477256 EAT45861.1 GFDL01002933 JAV32112.1 JXUM01121273 KQ566481 KXJ70098.1 GECU01009850 JAS97856.1 AE014297 AGB96086.1 AGB96087.1 GECZ01005529 JAS64240.1 UFQT01000466 SSX24603.1 GECU01028053 JAS79653.1 JRES01000835 KNC27977.1 GDHF01028106 GDHF01008414 GDHF01007329 JAI24208.1 JAI43900.1 JAI44985.1 GAMC01021191 GAMC01021190 GAMC01021189 GAMC01021188 GAMC01021187 JAB85366.1 KQ971354 EFA06980.2 KYB26448.1 GBXI01017552 JAC96739.1 GAKP01019065 GAKP01019064 JAC39887.1 KA650008 AFP64637.1 AJWK01011027 AJWK01011028 AJWK01011029 AJWK01011030 CH964232 EDW80561.1 GEZM01036980 GEZM01036979 JAV82300.1 CH954181 EDV48614.1 CH933806 EDW14453.2 CH916369 EDV93310.1 CH479179 EDW24790.1 GFDF01007015 JAV07069.1 GFDF01007014 JAV07070.1 AY118312 AAF55536.4 AAM48341.1 AGB96085.1 CM000364 EDX12460.1 OUUW01000013 SPP88021.1 CH940652 EDW59366.2 CH480827 EDW44622.1 GEZM01036983 GEZM01036982 JAV82294.1 CM000160 EDW95891.2 CM000070 EDY67948.2 CH902617 EDV42688.2 GGFK01005124 MBW38445.1 AXCM01006825 KYB26446.1 GEZM01036985 GEZM01036984 JAV82292.1 KC896834 AGY49605.1 AAAB01008846 EAA06222.6 ADMH02001467 ETN62472.1 APCN01004039 GGFJ01001662 MBW50803.1 GEZM01036981 GEZM01036978 JAV82301.1 ADTU01009967 GL888529 EGI59741.1 KQ976455 KYM85112.1 CCAG010019856 KQ981305 KYN42932.1 KB632313 ERL92165.1 GBHO01020581 GDHC01016161 JAG23023.1 JAQ02468.1 ATLV01013420 KE524855 KFB37945.1 AXCN02001220 NEVH01019377 PNF22567.1 GL446764 EFN87279.1 KK855460 PTY22675.1 KZ288325 PBC28034.1 LBMM01001175 KMQ96780.1 KQ435747 KOX76512.1 KQ759883 OAD62173.1 JXJN01015362 JXJN01015363 KQ982686 KYQ52327.1 APGK01051641 KB741198 ENN72969.1 KQ980734 KYN13796.1 KQ977276 KYN04467.1 KQ434896 KZC10786.1 KK107796 QOIP01000013 EZA47622.1 RLU15226.1 KQ414786 KOC60946.1 PYGN01000262 PSN49780.1 GBBI01002692 JAC16020.1 NNAY01004210 OXU18195.1
KPJ17062.1 KQ459604 KPI92140.1 AGBW02014918 OWR40862.1 GAIX01007184 JAA85376.1 GFDL01011126 JAV23919.1 GFDL01010729 JAV24316.1 DS231939 EDS27946.1 CH477256 EAT45861.1 GFDL01002933 JAV32112.1 JXUM01121273 KQ566481 KXJ70098.1 GECU01009850 JAS97856.1 AE014297 AGB96086.1 AGB96087.1 GECZ01005529 JAS64240.1 UFQT01000466 SSX24603.1 GECU01028053 JAS79653.1 JRES01000835 KNC27977.1 GDHF01028106 GDHF01008414 GDHF01007329 JAI24208.1 JAI43900.1 JAI44985.1 GAMC01021191 GAMC01021190 GAMC01021189 GAMC01021188 GAMC01021187 JAB85366.1 KQ971354 EFA06980.2 KYB26448.1 GBXI01017552 JAC96739.1 GAKP01019065 GAKP01019064 JAC39887.1 KA650008 AFP64637.1 AJWK01011027 AJWK01011028 AJWK01011029 AJWK01011030 CH964232 EDW80561.1 GEZM01036980 GEZM01036979 JAV82300.1 CH954181 EDV48614.1 CH933806 EDW14453.2 CH916369 EDV93310.1 CH479179 EDW24790.1 GFDF01007015 JAV07069.1 GFDF01007014 JAV07070.1 AY118312 AAF55536.4 AAM48341.1 AGB96085.1 CM000364 EDX12460.1 OUUW01000013 SPP88021.1 CH940652 EDW59366.2 CH480827 EDW44622.1 GEZM01036983 GEZM01036982 JAV82294.1 CM000160 EDW95891.2 CM000070 EDY67948.2 CH902617 EDV42688.2 GGFK01005124 MBW38445.1 AXCM01006825 KYB26446.1 GEZM01036985 GEZM01036984 JAV82292.1 KC896834 AGY49605.1 AAAB01008846 EAA06222.6 ADMH02001467 ETN62472.1 APCN01004039 GGFJ01001662 MBW50803.1 GEZM01036981 GEZM01036978 JAV82301.1 ADTU01009967 GL888529 EGI59741.1 KQ976455 KYM85112.1 CCAG010019856 KQ981305 KYN42932.1 KB632313 ERL92165.1 GBHO01020581 GDHC01016161 JAG23023.1 JAQ02468.1 ATLV01013420 KE524855 KFB37945.1 AXCN02001220 NEVH01019377 PNF22567.1 GL446764 EFN87279.1 KK855460 PTY22675.1 KZ288325 PBC28034.1 LBMM01001175 KMQ96780.1 KQ435747 KOX76512.1 KQ759883 OAD62173.1 JXJN01015362 JXJN01015363 KQ982686 KYQ52327.1 APGK01051641 KB741198 ENN72969.1 KQ980734 KYN13796.1 KQ977276 KYN04467.1 KQ434896 KZC10786.1 KK107796 QOIP01000013 EZA47622.1 RLU15226.1 KQ414786 KOC60946.1 PYGN01000262 PSN49780.1 GBBI01002692 JAC16020.1 NNAY01004210 OXU18195.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000007151
UP000002320
UP000008820
+ More
UP000069940 UP000249989 UP000000803 UP000037069 UP000007266 UP000075903 UP000095300 UP000075901 UP000092461 UP000007798 UP000008711 UP000009192 UP000001070 UP000095301 UP000008744 UP000192221 UP000000304 UP000268350 UP000008792 UP000001292 UP000002282 UP000001819 UP000007801 UP000075885 UP000075883 UP000075884 UP000007062 UP000000673 UP000075840 UP000076408 UP000069272 UP000005205 UP000007755 UP000078540 UP000078200 UP000075881 UP000092444 UP000078541 UP000030742 UP000030765 UP000075886 UP000075880 UP000235965 UP000008237 UP000242457 UP000036403 UP000053105 UP000005203 UP000092460 UP000075809 UP000019118 UP000078492 UP000078542 UP000076502 UP000053097 UP000279307 UP000053825 UP000092443 UP000245037 UP000192223 UP000091820 UP000215335
UP000069940 UP000249989 UP000000803 UP000037069 UP000007266 UP000075903 UP000095300 UP000075901 UP000092461 UP000007798 UP000008711 UP000009192 UP000001070 UP000095301 UP000008744 UP000192221 UP000000304 UP000268350 UP000008792 UP000001292 UP000002282 UP000001819 UP000007801 UP000075885 UP000075883 UP000075884 UP000007062 UP000000673 UP000075840 UP000076408 UP000069272 UP000005205 UP000007755 UP000078540 UP000078200 UP000075881 UP000092444 UP000078541 UP000030742 UP000030765 UP000075886 UP000075880 UP000235965 UP000008237 UP000242457 UP000036403 UP000053105 UP000005203 UP000092460 UP000075809 UP000019118 UP000078492 UP000078542 UP000076502 UP000053097 UP000279307 UP000053825 UP000092443 UP000245037 UP000192223 UP000091820 UP000215335
Interpro
SUPFAM
SSF56112
SSF56112
Gene 3D
CDD
ProteinModelPortal
H9IU60
A0A2H1X0J4
A0A2W1BP91
A0A194RHM1
A0A194PM08
A0A212EHA8
+ More
S4P1R1 A0A1Q3F8R2 A0A1Q3F9U0 B0WHS9 Q17GS6 A0A1S4F342 A0A1Q3FXD5 A0A182H9Q1 A0A1B6JF00 A0A0B4KGF0 A0A1B6GPA7 A0A336M6P4 A0A1B6HYA8 A0A0L0C6W2 A0A0K8UC92 W8AX18 D6WQI6 A0A182V8R4 A0A139WEP4 A0A0A1WE52 A0A1I8NM65 A0A034VCP3 T1PPM3 A0A182SIT0 A0A1B0CGD3 B4N9C8 A0A1Y1MCH3 B3NZ72 B4K940 B4JF29 A0A1I8MGR5 B4G5C4 A0A1W4U8S3 A0A1L8DKS6 A0A1L8DL51 Q9VE91 A0A0B4KHC3 B4QUG0 A0A3B0KS02 B4M405 B4IB64 A0A1Y1M9B7 B4PLA9 B5DY27 B3LWQ0 A0A2M4ACC3 A0A182PTP1 A0A182M5B1 A0A139WES9 A0A1Y1M919 A0A182N023 U5Q3U4 A0A340TB55 Q7PS40 W5JDN3 A0A182HIP5 A0A182YCM5 A0A2M4BCQ5 A0A182FJE3 A0A1Y1M9N6 A0A158NA49 F4X1E6 A0A195BK94 A0A1A9VEP9 A0A182KA76 A0A1B0F9Q4 A0A195FRL9 U4UQ62 A0A0A9XVX8 A0A084VJ01 A0A182QBL7 A0A182J288 A0A2J7Q1W3 E2BAK2 A0A2R7WVP8 A0A2A3E987 A0A0J7NWI4 A0A0N0BHP1 A0A310SWG2 A0A088A8E8 A0A1B0BJC4 A0A151WX40 N6SYX1 A0A195DLS6 A0A195CVA2 A0A154PFX7 A0A026VV67 A0A0L7QR25 A0A1A9XZB5 A0A2P8YZX4 A0A1W4WN85 A0A1A9W3J6 A0A023F3P7 A0A232EIM1
S4P1R1 A0A1Q3F8R2 A0A1Q3F9U0 B0WHS9 Q17GS6 A0A1S4F342 A0A1Q3FXD5 A0A182H9Q1 A0A1B6JF00 A0A0B4KGF0 A0A1B6GPA7 A0A336M6P4 A0A1B6HYA8 A0A0L0C6W2 A0A0K8UC92 W8AX18 D6WQI6 A0A182V8R4 A0A139WEP4 A0A0A1WE52 A0A1I8NM65 A0A034VCP3 T1PPM3 A0A182SIT0 A0A1B0CGD3 B4N9C8 A0A1Y1MCH3 B3NZ72 B4K940 B4JF29 A0A1I8MGR5 B4G5C4 A0A1W4U8S3 A0A1L8DKS6 A0A1L8DL51 Q9VE91 A0A0B4KHC3 B4QUG0 A0A3B0KS02 B4M405 B4IB64 A0A1Y1M9B7 B4PLA9 B5DY27 B3LWQ0 A0A2M4ACC3 A0A182PTP1 A0A182M5B1 A0A139WES9 A0A1Y1M919 A0A182N023 U5Q3U4 A0A340TB55 Q7PS40 W5JDN3 A0A182HIP5 A0A182YCM5 A0A2M4BCQ5 A0A182FJE3 A0A1Y1M9N6 A0A158NA49 F4X1E6 A0A195BK94 A0A1A9VEP9 A0A182KA76 A0A1B0F9Q4 A0A195FRL9 U4UQ62 A0A0A9XVX8 A0A084VJ01 A0A182QBL7 A0A182J288 A0A2J7Q1W3 E2BAK2 A0A2R7WVP8 A0A2A3E987 A0A0J7NWI4 A0A0N0BHP1 A0A310SWG2 A0A088A8E8 A0A1B0BJC4 A0A151WX40 N6SYX1 A0A195DLS6 A0A195CVA2 A0A154PFX7 A0A026VV67 A0A0L7QR25 A0A1A9XZB5 A0A2P8YZX4 A0A1W4WN85 A0A1A9W3J6 A0A023F3P7 A0A232EIM1
PDB
3PFQ
E-value=2.54217e-08,
Score=134
Ontologies
GO
PANTHER
Topology
Subcellular location
Cytoplasm
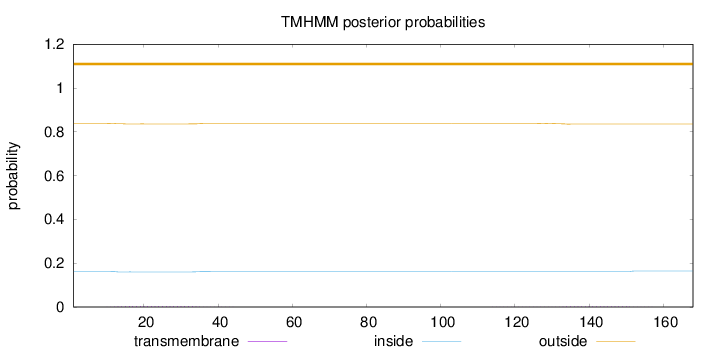
Length:
168
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.1475
Exp number, first 60 AAs:
0.09119
Total prob of N-in:
0.16193
outside
1 - 168
Population Genetic Test Statistics
Pi
313.915291
Theta
199.277193
Tajima's D
2.10349
CLR
0
CSRT
0.901054947252637
Interpretation
Uncertain
