Gene
KWMTBOMO14918 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA000774
Annotation
carboxyl/cholinesterase_6_precursor_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 2.134
Sequence
CDS
ATGATAAGACTGGGTCTGGTGATGCTGTTCCTAATACCTGGAACAGAACAGAATGTAATCAGTATAGCTCAGGGGAAGATACGGGGACGCGTCAGAGATGGTTACGTGACGTATGGCGGTATACCTTATGGATCTCTGATGCTTAAGGGGAGATTTAAGCAAGCCGGCATAGCTCCCGTTTGGCCTAACATACGAGAGTTCCACGAATCTCGCTGTTCAACAACTTCCGCCGCCGAGGAGTGTCTGCAGCTCGACGTCAACGTGCCCACGTCGGCTGCTCCAACGTGGCCTGTACTAGTCTGGGTCACAGGTTCAGACGGAAGTTATCACTCTGGACAACTGGTCAAGAAGGGAGTTATTGTAGTCATTGTGCGTCACAGGTTAGGCGCCCCTGGATTCTTATGCTACGAAAGAGATATACCAGGAAACGCAGGCGCAAAAGACGTTATAGCAGCATTACACTGGGTCAGAGACAACATAATTGCTTTCAAAGGAAACCCGGCAAGAGTTGTCGTCGCGGGCCAGGGTTTTGGAGCTGCCATGGTGGAAGCATTAACACTTACCCCAATGGCTGAAGGACTGTTCCACGGTGTTATATTGCAAAGCGGCAGCATATTAGCTCCGTGGGCTTTCAACTATGATGCCAAAGACAGGGCCAAAGTTATCGGAGAGCGTTTAGACAAAAAACGAGATTATGTCGATGTTTTTTTAAATGCCACAATGGAAGAATTATTTATTAAATCAAGTGAATCGAACACGCCATATTTCTCTTTTGGCCTTTGTCTGGAAGAACCACTGAAGAATGAGGAGCGGTTTTTCTTGGAGACGCCTTACGATTTGTTCGTGACAAAGAAGTCTAAATCTGTTCCAATGATAATCGGCTACAATAGTGATGAAGCGTATATATTCTCATCACTACTAAAGGAGTTTAAGGTTATTAATAAATTAACTAAAGATATTAGCTTTTTACTGCCATACGAGTTGCAATTTTTAAATAGTAGAGAAATGAATCAAGTTTCTCGACAAATCGAGGACATTTACTTCAGAAGAAATCGTACAATGACTGCTCTGCTGGCTTACCACAGGGACGTATACTTTTTAAGCCATGTTTATCGAAGTGCCAGTTATCACGCCGCGGTGTCATCCCTTGTCTACTTCTACACGTTTTCACACCTTGGAGAAGTCGGCGTTCAAAAAGAACCTGGAATCCAGAAAAGCGGAGCCGCTCATTCAGATGAACTTGCATATCTATTTTCTGGGCGAAATCTGGATAAAGAAGACGGCCTTGTCCAAGACCTTTTGGTACGATTTTGGGTCAACTTCGTGATACATTTAAATCCAACTCACCGACAACCACTTGACTGGGATCCAATGAATTACGAAAACCCTCGCCTACTCGATATAGGCGTTGAACCTAAGATGATCGACTACCCTTACGAGAAGACTTCAAAATTTTGGGAAGAAATTTATGACAATTCAGTTACCCTGATTTTAATTGGTTCAGCGATATATGCAGAGTCCTTCACTAAAAAATGTGATGTGCTGGTGAAGTTAGACTCGGGACCTGTGTGCGGGAGAGAGGAGAGCGCAAACAAAAATACCAAATACTTCAGTTTCCAGGGCATTCCGTATGCGAAGCCACCCGTAGGCGCCAGAAGATTTTCTGAACTGGAGCCACTTGAGCCATGGTCTGAACCGTTCTACGCTTACGAAGAAGGGCCGGCCTGTCCGTCGAGAGATATTACTTACGGCAGTATTACCGTCAAACGCAAAGGAATGAGTGAAAACTGTATTTATGCTAATGTATTTGTACCGGCGTCTGCGACTCTGAACAGCGATGAACTTTGCGAAGACAATTCACTGCCCATATTGGTGAATATCCACGGTGGTGGGTTCCAAACAGGATCTGGCAATAGAGATTTGCATGGACCGGAATTGTTGATGTTAAAAGATGTCATTGTAGTAAATTTTAATTACAGATTGGCCATTTTCGGATACTTGTCTCTTGCTTCTCATAAGATTCCTGGTAATAATGGACTGCGAGACATGGTGACGCTGCTAAAGTGGGTGCAGAGAAATGCAAAAGTATTCGGGGGAGACCCTAAGCGAGTGACAATACTCGGAGAAAGTGCTGGAGCAGCATCAGTACACCTTCTCATGTTGTCTCGGGCCTCCAAAGGACTATTCAACAAAGCTATTATAATGAGTGGTACCGCTATACCAAACTTCTATAGCTCGTCCCCAATATATGCCAAATACATCTCCGACATGTTTCTCGGAGAACTAGGTTTAAACTCTACCGAGTTAACGTCTGATGAAATTCATCAGACTTTAACTGAATTACCGTTAGATGATATTATGAGAGCCAATGACGTTGTGCAATACAGAGCGGGTTTAACTTCGTTCGTTCCGGTTGTAGAGAAAGAGGGTCATGATTATACCAGGATCATTGACGATGATCCGATTACTTTAATTAATCAAGGTGTGGGCAAGGACATTCCCTTACTAATGGGTTTCAATAAGGATGAAGGTGAATATTTCAAATGGATAATCACTATTTACAATATTGTGGACCGTTATAAAAGTAATCCGGCAGTTATTTTATCGCCAAGACTGGCGTACGAGTTGCCCCCACAAGAAGCTTCAGCCAAAGGAATATTCGTGGGGAAAAGATATTTTGACGGTGAGCCCACAGTGGATGGATTCGTAAAAAGCGTGACGGATATTTACTTTCAATATCCTATGATCAAACTGGCACAATGGAGAATACTTTTGAATTCGGCGCCGACATATTTTTACGAATTTTCGTATGAAAGCGACTTCAGCATTACAAAGAGAGCCAACTGGTTAAGCTACAAAGGAACTGCTCATGTCGAGGATCTGACGTACGTGTTCCACACTACCACGTTTCTCGGAAGTCAAGTGTCAGTGCCTCCTAAAACCAGAGATGACCAAATGAGGGATTGGATGTCTACTTTATTCAGTAACTACGTAAGATGCAATAATCCTACATGCAATAAGCGTGATGATCCTCGGTGGCCTCCCATTAACGAAGAAGAGTTAATGTTTCAAGTTATCAAAGAGCCCAATGTTTATAAGATGTCTTCATTACCAAAGCAGTTGGAGGACATGGTGGAATTCCATGACGGTGTAGACGATCAAGTCAGGATGCAGTCAGAAATCCTTAAGGAGCAAAAAAAGAAACATAGTATGTACCAGTAA
Protein
MIRLGLVMLFLIPGTEQNVISIAQGKIRGRVRDGYVTYGGIPYGSLMLKGRFKQAGIAPVWPNIREFHESRCSTTSAAEECLQLDVNVPTSAAPTWPVLVWVTGSDGSYHSGQLVKKGVIVVIVRHRLGAPGFLCYERDIPGNAGAKDVIAALHWVRDNIIAFKGNPARVVVAGQGFGAAMVEALTLTPMAEGLFHGVILQSGSILAPWAFNYDAKDRAKVIGERLDKKRDYVDVFLNATMEELFIKSSESNTPYFSFGLCLEEPLKNEERFFLETPYDLFVTKKSKSVPMIIGYNSDEAYIFSSLLKEFKVINKLTKDISFLLPYELQFLNSREMNQVSRQIEDIYFRRNRTMTALLAYHRDVYFLSHVYRSASYHAAVSSLVYFYTFSHLGEVGVQKEPGIQKSGAAHSDELAYLFSGRNLDKEDGLVQDLLVRFWVNFVIHLNPTHRQPLDWDPMNYENPRLLDIGVEPKMIDYPYEKTSKFWEEIYDNSVTLILIGSAIYAESFTKKCDVLVKLDSGPVCGREESANKNTKYFSFQGIPYAKPPVGARRFSELEPLEPWSEPFYAYEEGPACPSRDITYGSITVKRKGMSENCIYANVFVPASATLNSDELCEDNSLPILVNIHGGGFQTGSGNRDLHGPELLMLKDVIVVNFNYRLAIFGYLSLASHKIPGNNGLRDMVTLLKWVQRNAKVFGGDPKRVTILGESAGAASVHLLMLSRASKGLFNKAIIMSGTAIPNFYSSSPIYAKYISDMFLGELGLNSTELTSDEIHQTLTELPLDDIMRANDVVQYRAGLTSFVPVVEKEGHDYTRIIDDDPITLINQGVGKDIPLLMGFNKDEGEYFKWIITIYNIVDRYKSNPAVILSPRLAYELPPQEASAKGIFVGKRYFDGEPTVDGFVKSVTDIYFQYPMIKLAQWRILLNSAPTYFYEFSYESDFSITKRANWLSYKGTAHVEDLTYVFHTTTFLGSQVSVPPKTRDDQMRDWMSTLFSNYVRCNNPTCNKRDDPRWPPINEEELMFQVIKEPNVYKMSSLPKQLEDMVEFHDGVDDQVRMQSEILKEQKKKHSMYQ
Summary
Similarity
Belongs to the type-B carboxylesterase/lipase family.
Uniprot
A0A212EHB0
A0A0L7LFI0
A0A1W4XHU4
A0A194Q3V9
A0A1W4WJL4
A0A084VDQ2
+ More
A0A182Y3T5 A0A139WDB1 A0A182PSI4 A0A182VZM5 A0A139WFF6 A0A182JU79 A0A182NRT3 A0A2A3E5C0 A0A0L7L5N8 A0A1I8NLU5 A0A1B0G6N0 A0A0L7KNG8 A0A182WIA7 A0A182K2F0 A0A2H1VCP1 A0A182X1A6 A0A182FFU4 A0A182K987 A0A194QY63 A0A182FHW9 A0A182N7N5 A0A194RIH0 A0A1B0G4Z5 A0A0L7L9I4 A0A2H1WPZ4 A0A195EV77 A0A2A4JAV9
A0A182Y3T5 A0A139WDB1 A0A182PSI4 A0A182VZM5 A0A139WFF6 A0A182JU79 A0A182NRT3 A0A2A3E5C0 A0A0L7L5N8 A0A1I8NLU5 A0A1B0G6N0 A0A0L7KNG8 A0A182WIA7 A0A182K2F0 A0A2H1VCP1 A0A182X1A6 A0A182FFU4 A0A182K987 A0A194QY63 A0A182FHW9 A0A182N7N5 A0A194RIH0 A0A1B0G4Z5 A0A0L7L9I4 A0A2H1WPZ4 A0A195EV77 A0A2A4JAV9
EMBL
AGBW02014918
OWR40860.1
JTDY01001282
KOB74303.1
KQ459586
KPI98095.1
+ More
ATLV01011649 ATLV01011650 KE524705 KFB36096.1 KQ971360 KYB25958.1 KQ971352 KYB26710.1 KZ288364 PBC26890.1 JTDY01002769 KOB70767.1 CCAG010009612 JTDY01007971 KOB64827.1 ODYU01001848 SOQ38618.1 KQ461154 KPJ08516.1 KQ460205 KPJ17125.1 CCAG010019794 JTDY01002125 KOB72049.1 ODYU01010136 SOQ55032.1 KQ981965 KYN31797.1 NWSH01002285 PCG68674.1
ATLV01011649 ATLV01011650 KE524705 KFB36096.1 KQ971360 KYB25958.1 KQ971352 KYB26710.1 KZ288364 PBC26890.1 JTDY01002769 KOB70767.1 CCAG010009612 JTDY01007971 KOB64827.1 ODYU01001848 SOQ38618.1 KQ461154 KPJ08516.1 KQ460205 KPJ17125.1 CCAG010019794 JTDY01002125 KOB72049.1 ODYU01010136 SOQ55032.1 KQ981965 KYN31797.1 NWSH01002285 PCG68674.1
Proteomes
Interpro
Gene 3D
ProteinModelPortal
A0A212EHB0
A0A0L7LFI0
A0A1W4XHU4
A0A194Q3V9
A0A1W4WJL4
A0A084VDQ2
+ More
A0A182Y3T5 A0A139WDB1 A0A182PSI4 A0A182VZM5 A0A139WFF6 A0A182JU79 A0A182NRT3 A0A2A3E5C0 A0A0L7L5N8 A0A1I8NLU5 A0A1B0G6N0 A0A0L7KNG8 A0A182WIA7 A0A182K2F0 A0A2H1VCP1 A0A182X1A6 A0A182FFU4 A0A182K987 A0A194QY63 A0A182FHW9 A0A182N7N5 A0A194RIH0 A0A1B0G4Z5 A0A0L7L9I4 A0A2H1WPZ4 A0A195EV77 A0A2A4JAV9
A0A182Y3T5 A0A139WDB1 A0A182PSI4 A0A182VZM5 A0A139WFF6 A0A182JU79 A0A182NRT3 A0A2A3E5C0 A0A0L7L5N8 A0A1I8NLU5 A0A1B0G6N0 A0A0L7KNG8 A0A182WIA7 A0A182K2F0 A0A2H1VCP1 A0A182X1A6 A0A182FFU4 A0A182K987 A0A194QY63 A0A182FHW9 A0A182N7N5 A0A194RIH0 A0A1B0G4Z5 A0A0L7L9I4 A0A2H1WPZ4 A0A195EV77 A0A2A4JAV9
PDB
2FJ0
E-value=1.04847e-128,
Score=1182
Ontologies
Topology
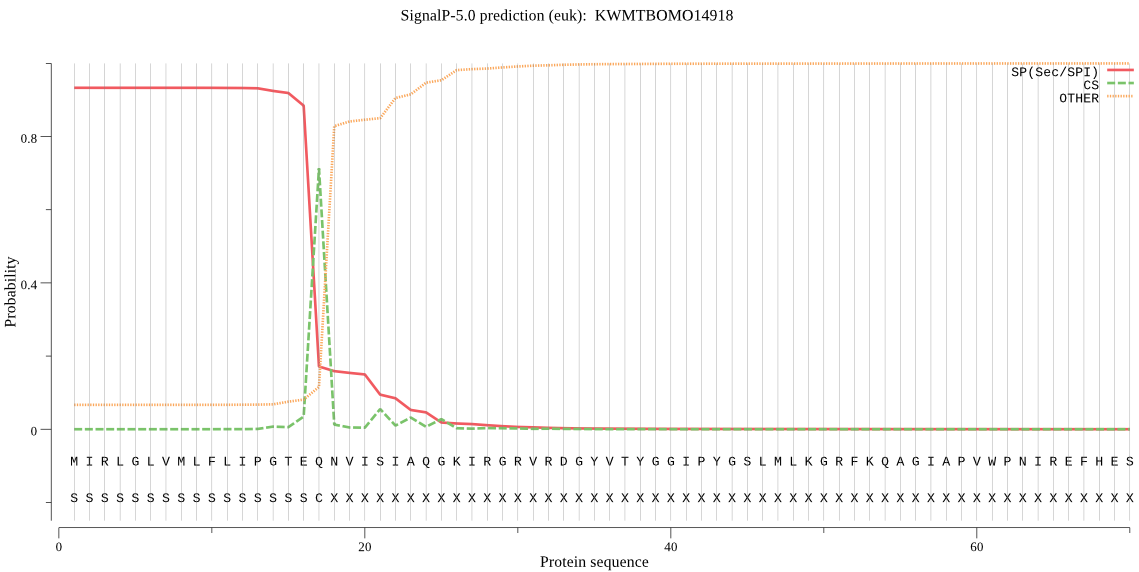
SignalP
Position: 1 - 17,
Likelihood: 0.933011
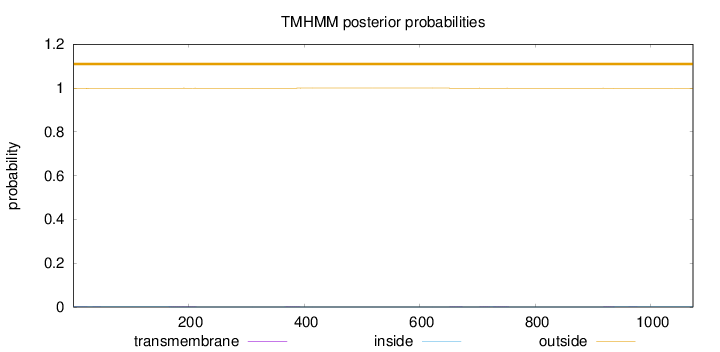
Length:
1073
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.1068
Exp number, first 60 AAs:
0.02084
Total prob of N-in:
0.00187
outside
1 - 1073
Population Genetic Test Statistics
Pi
223.815181
Theta
170.664725
Tajima's D
1.109655
CLR
0.169576
CSRT
0.687215639218039
Interpretation
Uncertain
