Pre Gene Modal
BGIBMGA000786
Annotation
PREDICTED:_ADP-ribosylation_factor-like_protein_2_[Plutella_xylostella]
Full name
ADP-ribosylation factor-like protein 2
Location in the cell
Cytoplasmic Reliability : 1.175 Mitochondrial Reliability : 1.683
Sequence
CDS
ATGGGTTTCTTGACAATTCTAAAGAAGCTGCGGCAGAAAGAGAAGGAAATGCGAATACTTATGTTAGGCCTAGACAATGCAGGGAAGACAACAATTTTGAAACGATTTAATGGTGAACCAATTGACACTATTTCTCCAACACTAGGCTTCAACATAAAAACTTTAGAACATAAGGGATTTAAGTTGAACATTTGGGATGTTGGGGGACAAAAGTCTTTGAGATCATACTGGAAGAACTACTTCGAGAGTACGGATGGAGTAGCGTGGGTGGTGGACGCGGCGGACGGGGCGCGACTGTCGGCGTGCGCGGCAGAACTCCGCGCGCTGCTCACGGAGGAGCGTCTGGCGGCCGCCACGCTGCTCGTGCTCGCCAACAAGTGCGACCTACCCGGAGCACTCTCCTTGCAACAAATACGAGAGGCTCTAGATCTCGACAGCATAAAATCTCATCACTGGCGCATCGTGAGGTGCTCTGCAGTCACCGGAGAGAACCTTTTAGAAGGAATCGACTGGATGTTAGACGATATTGCGTCGAGAATATTCACTCTAGACTGA
Protein
MGFLTILKKLRQKEKEMRILMLGLDNAGKTTILKRFNGEPIDTISPTLGFNIKTLEHKGFKLNIWDVGGQKSLRSYWKNYFESTDGVAWVVDAADGARLSACAAELRALLTEERLAAATLLVLANKCDLPGALSLQQIREALDLDSIKSHHWRIVRCSAVTGENLLEGIDWMLDDIASRIFTLD
Summary
Description
Small GTP-binding protein which cycles between an inactive GDP-bound and an active GTP-bound form, and the rate of cycling is regulated by guanine nucleotide exchange factors (GEF) and GTPase-activating proteins (GAP). GTP-binding protein that does not act as an allosteric activator of the cholera toxin catalytic subunit. Regulates formation of new microtubules and centrosome integrity. Prevents the TBCD-induced microtubule destruction. Participates in association with TBCD, in the disassembly of the apical junction complexes. Antagonizes the effect of TBCD on epithelial cell detachment and tight and adherens junctions disassembly. Together with ARL2, plays a role in the nuclear translocation, retention and transcriptional activity of STAT3. Component of a regulated secretory pathway involved in Ca(2+)-dependent release of acetylcholine. Required for normal progress through the cell cycle.
Small GTP-binding protein which cycles between an inactive GDP-bound and an active GTP-bound form, and the rate of cycling is regulated by guanine nucleotide exchange factors (GEF) and GTPase-activating proteins (GAP). GTP-binding protein that does not act as an allosteric activator of the cholera toxin catalytic subunit. Regulates formation of new microtubules and centrosome integrity. Prevents the TBCD-induced microtubule destruction. Participates in association with TBCD, in the disassembly of the apical junction complexes. Antagonizes the effect of TBCD on epithelial cell detachment and tight and adherens junctions disassembly. Together with ARL2, plays a role in the nuclear translocation, retention and transcriptional activity of STAT3. Component of a regulated secretory pathway involved in Ca(2+)-dependent release of acetylcholine. Required for normal progress through the cell cycle (By similarity).
Small GTP-binding protein which cycles between an inactive GDP-bound and an active GTP-bound form, and the rate of cycling is regulated by guanine nucleotide exchange factors (GEF) and GTPase-activating proteins (GAP). GTP-binding protein that does not act as an allosteric activator of the cholera toxin catalytic subunit. Regulates formation of new microtubules and centrosome integrity. Prevents the TBCD-induced microtubule destruction. Participates in association with TBCD, in the disassembly of the apical junction complexes. Antagonizes the effect of TBCD on epithelial cell detachment and tight and adherens junctions disassembly. Together with ARL2, plays a role in the nuclear translocation, retention and transcriptional activity of STAT3. Component of a regulated secretory pathway involved in Ca(2+)-dependent release of acetylcholine. Required for normal progress through the cell cycle (By similarity).
Subunit
Found in a complex with ARL2, ARL2BP and SLC25A6. Found in a complex with at least ARL2, PPP2CB, PPP2R1A, PPP2R2A, PPP2R5E and TBCD. Interacts with ELMOD2. The GTP-bound form interacts with ARL2BP. Interacts with TBCD; the GDP-bound form interacts preferentially with TBCD. Interacts with UNC119 (By similarity). Found in a complex with ARL2, ARL2BP and SLC25A4. The GTP-bound form interacts with PDE6D.
Found in a complex with ARL2, ARL2BP and SLC25A6. Found in a complex with at least ARL2, PPP2CB, PPP2R1A, PPP2R2A, PPP2R5E and TBCD. Found in a complex with ARL2, ARL2BP and SLC25A4. The GTP-bound form interacts with PDE6D. Interacts with ELMOD2. The GTP-bound form interacts with ARL2BP. Interacts, preferentially in its GDP-bound state, with TBCD. Interacts with UNC119.
Found in a complex with ARL2, ARL2BP and SLC25A6. Found in a complex with at least ARL2, PPP2CB, PPP2R1A, PPP2R2A, PPP2R5E and TBCD. Interacts with ELMOD2. The GTP-bound form interacts with ARL2BP. The GDP-bound form interacts preferentially with TBCD. Interacts with UNC119. Found in a complex with ARL2, ARL2BP and SLC25A4. The GTP-bound form interacts with PDE6D (By similarity).
Interacts with ELMOD2. Interacts with ARL2BP; the GTP-bound form interacts with ARL2BP. The GDP-bound form interacts preferentially with TBCD. Interacts with UNC119. Found in a complex with ARL2, ARL2BP and SLC25A4. The GTP-bound form interacts with PDE6D (By similarity). Found in a complex with ARL2, ARL2BP and SLC25A6. Found in a complex with at least ARL2, PPP2CB, PPP2R1A, PPP2R2A, PPP2R5E and TBCD.
Found in a complex with ARL2, ARL2BP and SLC25A6. Found in a complex with at least ARL2, PPP2CB, PPP2R1A, PPP2R2A, PPP2R5E and TBCD. Found in a complex with ARL2, ARL2BP and SLC25A4. The GTP-bound form interacts with PDE6D. Interacts with ELMOD2. The GTP-bound form interacts with ARL2BP. Interacts, preferentially in its GDP-bound state, with TBCD. Interacts with UNC119.
Found in a complex with ARL2, ARL2BP and SLC25A6. Found in a complex with at least ARL2, PPP2CB, PPP2R1A, PPP2R2A, PPP2R5E and TBCD. Interacts with ELMOD2. The GTP-bound form interacts with ARL2BP. The GDP-bound form interacts preferentially with TBCD. Interacts with UNC119. Found in a complex with ARL2, ARL2BP and SLC25A4. The GTP-bound form interacts with PDE6D (By similarity).
Interacts with ELMOD2. Interacts with ARL2BP; the GTP-bound form interacts with ARL2BP. The GDP-bound form interacts preferentially with TBCD. Interacts with UNC119. Found in a complex with ARL2, ARL2BP and SLC25A4. The GTP-bound form interacts with PDE6D (By similarity). Found in a complex with ARL2, ARL2BP and SLC25A6. Found in a complex with at least ARL2, PPP2CB, PPP2R1A, PPP2R2A, PPP2R5E and TBCD.
Similarity
Belongs to the small GTPase superfamily. Arf family.
Keywords
3D-structure
Cell cycle
Complete proteome
Cytoplasm
Cytoskeleton
GTP-binding
Isopeptide bond
Lipoprotein
Mitochondrion
Myristate
Nucleotide-binding
Nucleus
Phosphoprotein
Reference proteome
Ubl conjugation
Alternative splicing
Polymorphism
Feature
chain ADP-ribosylation factor-like protein 2
splice variant In isoform 2.
sequence variant In dbSNP:rs664226.
splice variant In isoform 2.
sequence variant In dbSNP:rs664226.
Uniprot
A0A194RGX5
I4DP88
A0A2H1W2R7
A0A212EGZ5
A0A2W1BT10
A0A2A4J391
+ More
A0A1E1W6W8 J3JY06 H9IU56 A0A3S0ZJG6 A0A0B6ZUQ0 A0A2U9BAA7 A0A2C9JEU8 G3Q8H1 G1C2I8 I3K7D0 A0A3P8QR07 A0A3P9B335 A0A3Q0QTC2 K1RUY4 A0A3Q3R5E9 N6TNK7 I3N6L1 A0A3Q1AKG1 A0A3P8TXC6 F6RR25 A0A3B5BAX7 A0A1U8DFC6 A0A3Q1EQ76 A0A3Q1INZ5 G3QRA0 A0A2R9C7Z2 A0A2K6FWW4 K7CT71 A0A3B4WDX5 A0A3B4VGI2 A0A2K5WTW8 A0A0D9R5D2 A0A2K6NI28 A0A2K5L9F0 A0A2K5KFV8 H9ESA6 Q9D0J4 V3ZG06 A0A2K6AME1 A0A2U3Y0C8 A0A3B4GTJ1 L5KQZ6 A0A3Q3A154 A0A096N6Q0 A7STE6 A0A3Q3LF30 A0A3Q7TY33 H0XZF2 A0A1U7Q4W4 G1Q1M1 L5LEQ8 A0A3Q7R312 A0A2U3VGY0 A0A3Q7UFT0 Q53YD8 P36404 A0A1A8C0S3 O08697 K9IGY1 A0A2I4C3C6 L8IPX1 W5PP36 Q2TA37 G5B6M8 K7G987 A0A1J1HGL1 A0A341BTR7 A0A2U4CAZ7 A0A2Y9PV80 A0A2Y9FBX6 U3AVV9 A0A146NXT7 U3D220 A0A1A8R6R6 A0A1A8KGZ5 A0A1A8APT9 A0A0E9X524 A0A2Y9JP06 M3YM18 A0A340XL08 A0A2D4P442 A0A1A8LHQ7 A0A3P8WHU4 A0A3B5KNA9 C3Z2D6 A0A3P9Q8C1 A0A0S7H0T4 A0A3B3VKE9 A0A287B226 A0A2K6MKG5 A0A077W8T9 A0A2Y9GN60 A0A1S2ZFN0 A0A091DDS4
A0A1E1W6W8 J3JY06 H9IU56 A0A3S0ZJG6 A0A0B6ZUQ0 A0A2U9BAA7 A0A2C9JEU8 G3Q8H1 G1C2I8 I3K7D0 A0A3P8QR07 A0A3P9B335 A0A3Q0QTC2 K1RUY4 A0A3Q3R5E9 N6TNK7 I3N6L1 A0A3Q1AKG1 A0A3P8TXC6 F6RR25 A0A3B5BAX7 A0A1U8DFC6 A0A3Q1EQ76 A0A3Q1INZ5 G3QRA0 A0A2R9C7Z2 A0A2K6FWW4 K7CT71 A0A3B4WDX5 A0A3B4VGI2 A0A2K5WTW8 A0A0D9R5D2 A0A2K6NI28 A0A2K5L9F0 A0A2K5KFV8 H9ESA6 Q9D0J4 V3ZG06 A0A2K6AME1 A0A2U3Y0C8 A0A3B4GTJ1 L5KQZ6 A0A3Q3A154 A0A096N6Q0 A7STE6 A0A3Q3LF30 A0A3Q7TY33 H0XZF2 A0A1U7Q4W4 G1Q1M1 L5LEQ8 A0A3Q7R312 A0A2U3VGY0 A0A3Q7UFT0 Q53YD8 P36404 A0A1A8C0S3 O08697 K9IGY1 A0A2I4C3C6 L8IPX1 W5PP36 Q2TA37 G5B6M8 K7G987 A0A1J1HGL1 A0A341BTR7 A0A2U4CAZ7 A0A2Y9PV80 A0A2Y9FBX6 U3AVV9 A0A146NXT7 U3D220 A0A1A8R6R6 A0A1A8KGZ5 A0A1A8APT9 A0A0E9X524 A0A2Y9JP06 M3YM18 A0A340XL08 A0A2D4P442 A0A1A8LHQ7 A0A3P8WHU4 A0A3B5KNA9 C3Z2D6 A0A3P9Q8C1 A0A0S7H0T4 A0A3B3VKE9 A0A287B226 A0A2K6MKG5 A0A077W8T9 A0A2Y9GN60 A0A1S2ZFN0 A0A091DDS4
Pubmed
26354079
22651552
22118469
28756777
22516182
19121390
+ More
15562597 25186727 22992520 23537049 19892987 22398555 22722832 16136131 25362486 17431167 25319552 16141072 15489334 11809823 15979089 21183079 11980706 23254933 23258410 17615350 21993624 8125298 9373149 11181995 8415637 15146197 16554811 10488091 10831612 11303027 11847227 16525022 17452337 18588884 18234692 18981177 20740604 21269460 27666374 19368893 9208929 12527357 12912990 22673903 22751099 20809919 17704193 21993625 17381049 25243066 25613341 24487278 21551351 18563158 30723633
15562597 25186727 22992520 23537049 19892987 22398555 22722832 16136131 25362486 17431167 25319552 16141072 15489334 11809823 15979089 21183079 11980706 23254933 23258410 17615350 21993624 8125298 9373149 11181995 8415637 15146197 16554811 10488091 10831612 11303027 11847227 16525022 17452337 18588884 18234692 18981177 20740604 21269460 27666374 19368893 9208929 12527357 12912990 22673903 22751099 20809919 17704193 21993625 17381049 25243066 25613341 24487278 21551351 18563158 30723633
EMBL
KQ460205
KPJ17068.1
AK403472
KQ459604
BAM19728.1
KPI92147.1
+ More
ODYU01005693 SOQ46804.1 AGBW02014998 OWR40741.1 KZ149945 PZC76784.1 NWSH01003483 PCG66206.1 GDQN01008463 JAT82591.1 BT128129 AEE63090.1 BABH01034161 RQTK01000397 RUS80356.1 HACG01025448 CEK72313.1 CP026246 AWP00891.1 JF728807 AEJ38209.1 AERX01028350 JH817779 EKC38536.1 APGK01057871 KB741282 KB632233 ENN70840.1 ERL90418.1 AGTP01072426 CABD030079703 AJFE02101268 AACZ04016214 GABC01010578 GABF01004566 GABD01008716 GABE01008841 GABE01008840 JAA00760.1 JAA17579.1 JAA24384.1 JAA35898.1 AQIA01019877 AQIB01078739 JSUE03012071 JSUE03012072 JU321509 JU473700 JV045755 AFE65265.1 AFH30504.1 AFI35826.1 AF143680 AK011366 BC060259 KB202481 ESO90138.1 KB030625 ELK13371.1 AHZZ02006315 AHZZ02006316 DS469795 EDO32994.1 AAQR03147593 AAQR03147594 AAPE02049467 KB112964 ELK24366.1 BT006674 AK223502 CH471076 AAP35320.1 BAD97222.1 EAW74332.1 L13687 AF493888 CN338497 AP000436 BC002530 HADZ01009218 HAEA01012736 SBP73159.1 Y12708 GABZ01008152 JAA45373.1 JH880973 ELR57569.1 AMGL01061832 AMGL01061833 BC111133 JH168664 GEBF01006702 EHB04905.1 JAN96930.1 AGCU01174361 CVRI01000003 CRK87011.1 GAMT01007473 JAB04388.1 GCES01150452 JAQ35870.1 GAMT01007474 GAMT01007472 GAMR01001524 GAMR01001523 GAMR01001522 GAMQ01002843 JAB04387.1 JAB32408.1 JAB39008.1 HAEI01007238 HAEH01016435 SBS00944.1 HAED01006307 HAEE01011670 SBR31720.1 HADY01018213 HAEJ01016484 SBP56698.1 GBXM01010770 JAH97807.1 AEYP01074240 IACN01045812 LAB52741.1 HAEF01006750 HAEG01013983 SBR44132.1 GG666574 EEN53187.1 GBYX01446759 JAO34687.1 AEMK02000006 DQIR01149383 DQIR01295787 HDB04860.1 LK023313 CDS03456.1 KN122776 KFO28430.1
ODYU01005693 SOQ46804.1 AGBW02014998 OWR40741.1 KZ149945 PZC76784.1 NWSH01003483 PCG66206.1 GDQN01008463 JAT82591.1 BT128129 AEE63090.1 BABH01034161 RQTK01000397 RUS80356.1 HACG01025448 CEK72313.1 CP026246 AWP00891.1 JF728807 AEJ38209.1 AERX01028350 JH817779 EKC38536.1 APGK01057871 KB741282 KB632233 ENN70840.1 ERL90418.1 AGTP01072426 CABD030079703 AJFE02101268 AACZ04016214 GABC01010578 GABF01004566 GABD01008716 GABE01008841 GABE01008840 JAA00760.1 JAA17579.1 JAA24384.1 JAA35898.1 AQIA01019877 AQIB01078739 JSUE03012071 JSUE03012072 JU321509 JU473700 JV045755 AFE65265.1 AFH30504.1 AFI35826.1 AF143680 AK011366 BC060259 KB202481 ESO90138.1 KB030625 ELK13371.1 AHZZ02006315 AHZZ02006316 DS469795 EDO32994.1 AAQR03147593 AAQR03147594 AAPE02049467 KB112964 ELK24366.1 BT006674 AK223502 CH471076 AAP35320.1 BAD97222.1 EAW74332.1 L13687 AF493888 CN338497 AP000436 BC002530 HADZ01009218 HAEA01012736 SBP73159.1 Y12708 GABZ01008152 JAA45373.1 JH880973 ELR57569.1 AMGL01061832 AMGL01061833 BC111133 JH168664 GEBF01006702 EHB04905.1 JAN96930.1 AGCU01174361 CVRI01000003 CRK87011.1 GAMT01007473 JAB04388.1 GCES01150452 JAQ35870.1 GAMT01007474 GAMT01007472 GAMR01001524 GAMR01001523 GAMR01001522 GAMQ01002843 JAB04387.1 JAB32408.1 JAB39008.1 HAEI01007238 HAEH01016435 SBS00944.1 HAED01006307 HAEE01011670 SBR31720.1 HADY01018213 HAEJ01016484 SBP56698.1 GBXM01010770 JAH97807.1 AEYP01074240 IACN01045812 LAB52741.1 HAEF01006750 HAEG01013983 SBR44132.1 GG666574 EEN53187.1 GBYX01446759 JAO34687.1 AEMK02000006 DQIR01149383 DQIR01295787 HDB04860.1 LK023313 CDS03456.1 KN122776 KFO28430.1
Proteomes
UP000053240
UP000053268
UP000007151
UP000218220
UP000005204
UP000271974
+ More
UP000246464 UP000076420 UP000007635 UP000005207 UP000265100 UP000265160 UP000261340 UP000005408 UP000261600 UP000019118 UP000030742 UP000005215 UP000257160 UP000265080 UP000002281 UP000261400 UP000189705 UP000257200 UP000265040 UP000001519 UP000240080 UP000233160 UP000002277 UP000261360 UP000261420 UP000233100 UP000029965 UP000233200 UP000233060 UP000233080 UP000006718 UP000000589 UP000030746 UP000233120 UP000245341 UP000261460 UP000010552 UP000264800 UP000028761 UP000001593 UP000261640 UP000286640 UP000005225 UP000189706 UP000001074 UP000286641 UP000245340 UP000286642 UP000005640 UP000002494 UP000192220 UP000002356 UP000009136 UP000006813 UP000007267 UP000183832 UP000252040 UP000245320 UP000248483 UP000248484 UP000008225 UP000248482 UP000000715 UP000265300 UP000265120 UP000005226 UP000001554 UP000242638 UP000261500 UP000008227 UP000233180 UP000248481 UP000079721 UP000028990
UP000246464 UP000076420 UP000007635 UP000005207 UP000265100 UP000265160 UP000261340 UP000005408 UP000261600 UP000019118 UP000030742 UP000005215 UP000257160 UP000265080 UP000002281 UP000261400 UP000189705 UP000257200 UP000265040 UP000001519 UP000240080 UP000233160 UP000002277 UP000261360 UP000261420 UP000233100 UP000029965 UP000233200 UP000233060 UP000233080 UP000006718 UP000000589 UP000030746 UP000233120 UP000245341 UP000261460 UP000010552 UP000264800 UP000028761 UP000001593 UP000261640 UP000286640 UP000005225 UP000189706 UP000001074 UP000286641 UP000245340 UP000286642 UP000005640 UP000002494 UP000192220 UP000002356 UP000009136 UP000006813 UP000007267 UP000183832 UP000252040 UP000245320 UP000248483 UP000248484 UP000008225 UP000248482 UP000000715 UP000265300 UP000265120 UP000005226 UP000001554 UP000242638 UP000261500 UP000008227 UP000233180 UP000248481 UP000079721 UP000028990
Pfam
PF00025 Arf
SUPFAM
SSF52540
SSF52540
ProteinModelPortal
A0A194RGX5
I4DP88
A0A2H1W2R7
A0A212EGZ5
A0A2W1BT10
A0A2A4J391
+ More
A0A1E1W6W8 J3JY06 H9IU56 A0A3S0ZJG6 A0A0B6ZUQ0 A0A2U9BAA7 A0A2C9JEU8 G3Q8H1 G1C2I8 I3K7D0 A0A3P8QR07 A0A3P9B335 A0A3Q0QTC2 K1RUY4 A0A3Q3R5E9 N6TNK7 I3N6L1 A0A3Q1AKG1 A0A3P8TXC6 F6RR25 A0A3B5BAX7 A0A1U8DFC6 A0A3Q1EQ76 A0A3Q1INZ5 G3QRA0 A0A2R9C7Z2 A0A2K6FWW4 K7CT71 A0A3B4WDX5 A0A3B4VGI2 A0A2K5WTW8 A0A0D9R5D2 A0A2K6NI28 A0A2K5L9F0 A0A2K5KFV8 H9ESA6 Q9D0J4 V3ZG06 A0A2K6AME1 A0A2U3Y0C8 A0A3B4GTJ1 L5KQZ6 A0A3Q3A154 A0A096N6Q0 A7STE6 A0A3Q3LF30 A0A3Q7TY33 H0XZF2 A0A1U7Q4W4 G1Q1M1 L5LEQ8 A0A3Q7R312 A0A2U3VGY0 A0A3Q7UFT0 Q53YD8 P36404 A0A1A8C0S3 O08697 K9IGY1 A0A2I4C3C6 L8IPX1 W5PP36 Q2TA37 G5B6M8 K7G987 A0A1J1HGL1 A0A341BTR7 A0A2U4CAZ7 A0A2Y9PV80 A0A2Y9FBX6 U3AVV9 A0A146NXT7 U3D220 A0A1A8R6R6 A0A1A8KGZ5 A0A1A8APT9 A0A0E9X524 A0A2Y9JP06 M3YM18 A0A340XL08 A0A2D4P442 A0A1A8LHQ7 A0A3P8WHU4 A0A3B5KNA9 C3Z2D6 A0A3P9Q8C1 A0A0S7H0T4 A0A3B3VKE9 A0A287B226 A0A2K6MKG5 A0A077W8T9 A0A2Y9GN60 A0A1S2ZFN0 A0A091DDS4
A0A1E1W6W8 J3JY06 H9IU56 A0A3S0ZJG6 A0A0B6ZUQ0 A0A2U9BAA7 A0A2C9JEU8 G3Q8H1 G1C2I8 I3K7D0 A0A3P8QR07 A0A3P9B335 A0A3Q0QTC2 K1RUY4 A0A3Q3R5E9 N6TNK7 I3N6L1 A0A3Q1AKG1 A0A3P8TXC6 F6RR25 A0A3B5BAX7 A0A1U8DFC6 A0A3Q1EQ76 A0A3Q1INZ5 G3QRA0 A0A2R9C7Z2 A0A2K6FWW4 K7CT71 A0A3B4WDX5 A0A3B4VGI2 A0A2K5WTW8 A0A0D9R5D2 A0A2K6NI28 A0A2K5L9F0 A0A2K5KFV8 H9ESA6 Q9D0J4 V3ZG06 A0A2K6AME1 A0A2U3Y0C8 A0A3B4GTJ1 L5KQZ6 A0A3Q3A154 A0A096N6Q0 A7STE6 A0A3Q3LF30 A0A3Q7TY33 H0XZF2 A0A1U7Q4W4 G1Q1M1 L5LEQ8 A0A3Q7R312 A0A2U3VGY0 A0A3Q7UFT0 Q53YD8 P36404 A0A1A8C0S3 O08697 K9IGY1 A0A2I4C3C6 L8IPX1 W5PP36 Q2TA37 G5B6M8 K7G987 A0A1J1HGL1 A0A341BTR7 A0A2U4CAZ7 A0A2Y9PV80 A0A2Y9FBX6 U3AVV9 A0A146NXT7 U3D220 A0A1A8R6R6 A0A1A8KGZ5 A0A1A8APT9 A0A0E9X524 A0A2Y9JP06 M3YM18 A0A340XL08 A0A2D4P442 A0A1A8LHQ7 A0A3P8WHU4 A0A3B5KNA9 C3Z2D6 A0A3P9Q8C1 A0A0S7H0T4 A0A3B3VKE9 A0A287B226 A0A2K6MKG5 A0A077W8T9 A0A2Y9GN60 A0A1S2ZFN0 A0A091DDS4
PDB
1KSJ
E-value=3.09289e-63,
Score=609
Ontologies
GO
GO:0005525
GO:0005737
GO:0019003
GO:0031116
GO:0015630
GO:0005925
GO:0003924
GO:0005829
GO:0051457
GO:0005730
GO:0005813
GO:0007098
GO:0005794
GO:0005758
GO:0034260
GO:0015870
GO:0031113
GO:0070830
GO:0016328
GO:0010811
GO:0005634
GO:0050796
GO:0005095
GO:0007021
GO:0005759
GO:0034333
GO:0005739
GO:0005622
GO:0007264
GO:0006468
GO:0004672
GO:0004697
GO:0007154
GO:0016020
GO:0005112
GO:0015930
Topology
Subcellular location
Mitochondrion intermembrane space
The complex formed with ARL2BP, ARL2 and SLC25A6 is expressed in mitochondria. Not detected in the Golgi, nucleus and on the mitotic spindle. Centrosome-associated throughout the cell cycle. Not detected to interphase microtubules (By similarity). The complex formed with ARL2BP, ARL2 and SLC25A4 is expressed in mitochondria. With evidence from 4 publications.
Cytoplasm The complex formed with ARL2BP, ARL2 and SLC25A6 is expressed in mitochondria. Not detected in the Golgi, nucleus and on the mitotic spindle. Centrosome-associated throughout the cell cycle. Not detected to interphase microtubules (By similarity). The complex formed with ARL2BP, ARL2 and SLC25A4 is expressed in mitochondria. With evidence from 4 publications.
Cytoskeleton The complex formed with ARL2BP, ARL2 and SLC25A6 is expressed in mitochondria. Not detected in the Golgi, nucleus and on the mitotic spindle. Centrosome-associated throughout the cell cycle. Not detected to interphase microtubules (By similarity). The complex formed with ARL2BP, ARL2 and SLC25A4 is expressed in mitochondria. With evidence from 4 publications.
Microtubule organizing center The complex formed with ARL2BP, ARL2 and SLC25A6 is expressed in mitochondria. Not detected in the Golgi, nucleus and on the mitotic spindle. Centrosome-associated throughout the cell cycle. Not detected to interphase microtubules (By similarity). The complex formed with ARL2BP, ARL2 and SLC25A4 is expressed in mitochondria. With evidence from 4 publications.
Centrosome The complex formed with ARL2BP, ARL2 and SLC25A6 is expressed in mitochondria. Not detected in the Golgi, nucleus and on the mitotic spindle. Centrosome-associated throughout the cell cycle. Not detected to interphase microtubules (By similarity). The complex formed with ARL2BP, ARL2 and SLC25A4 is expressed in mitochondria. With evidence from 4 publications.
Nucleus The complex formed with ARL2BP, ARL2 and SLC25A6 is expressed in mitochondria. Not detected in the Golgi, nucleus and on the mitotic spindle. Centrosome-associated throughout the cell cycle. Not detected to interphase microtubules (By similarity). The complex formed with ARL2BP, ARL2 and SLC25A4 is expressed in mitochondria. With evidence from 4 publications.
Mitochondrion The complex formed with ARL2BP, ARL2 and SLC25A4 is expressed in mitochondria. Not detected in the Golgi, nucleus and on the mitotic spindle. Centrosome-associated throughout the cell cycle. Not detected to interphase microtubules (By similarity). The complex formed with ARL2BP, ARL2 and SLC25A6 is expressed in mitochondria. With evidence from 1 publications.
Cytoplasm The complex formed with ARL2BP, ARL2 and SLC25A6 is expressed in mitochondria. Not detected in the Golgi, nucleus and on the mitotic spindle. Centrosome-associated throughout the cell cycle. Not detected to interphase microtubules (By similarity). The complex formed with ARL2BP, ARL2 and SLC25A4 is expressed in mitochondria. With evidence from 4 publications.
Cytoskeleton The complex formed with ARL2BP, ARL2 and SLC25A6 is expressed in mitochondria. Not detected in the Golgi, nucleus and on the mitotic spindle. Centrosome-associated throughout the cell cycle. Not detected to interphase microtubules (By similarity). The complex formed with ARL2BP, ARL2 and SLC25A4 is expressed in mitochondria. With evidence from 4 publications.
Microtubule organizing center The complex formed with ARL2BP, ARL2 and SLC25A6 is expressed in mitochondria. Not detected in the Golgi, nucleus and on the mitotic spindle. Centrosome-associated throughout the cell cycle. Not detected to interphase microtubules (By similarity). The complex formed with ARL2BP, ARL2 and SLC25A4 is expressed in mitochondria. With evidence from 4 publications.
Centrosome The complex formed with ARL2BP, ARL2 and SLC25A6 is expressed in mitochondria. Not detected in the Golgi, nucleus and on the mitotic spindle. Centrosome-associated throughout the cell cycle. Not detected to interphase microtubules (By similarity). The complex formed with ARL2BP, ARL2 and SLC25A4 is expressed in mitochondria. With evidence from 4 publications.
Nucleus The complex formed with ARL2BP, ARL2 and SLC25A6 is expressed in mitochondria. Not detected in the Golgi, nucleus and on the mitotic spindle. Centrosome-associated throughout the cell cycle. Not detected to interphase microtubules (By similarity). The complex formed with ARL2BP, ARL2 and SLC25A4 is expressed in mitochondria. With evidence from 4 publications.
Mitochondrion The complex formed with ARL2BP, ARL2 and SLC25A4 is expressed in mitochondria. Not detected in the Golgi, nucleus and on the mitotic spindle. Centrosome-associated throughout the cell cycle. Not detected to interphase microtubules (By similarity). The complex formed with ARL2BP, ARL2 and SLC25A6 is expressed in mitochondria. With evidence from 1 publications.
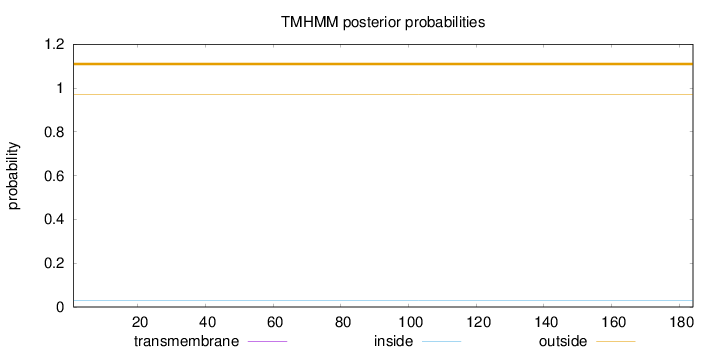
Length:
184
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00083
Exp number, first 60 AAs:
0
Total prob of N-in:
0.02901
outside
1 - 184
Population Genetic Test Statistics
Pi
281.920175
Theta
184.896105
Tajima's D
1.735196
CLR
0.001963
CSRT
0.837458127093645
Interpretation
Uncertain
