Pre Gene Modal
BGIBMGA014453
Annotation
aldo-keto_reductase_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 3.679
Sequence
CDS
ATGGCCGCGCCAGTCCCCGTCATATTCAACAACGGACGCACGTGTCCCGTCATCGGACTAGGAACTTGGAAGTCTAAGCCAGGAGAGGTGACCCAAGCAGTCAAAGACGCGATCGATATCGGCTACCGGCACGTGGACTGCGCCCACATCTACCTCAATGAGAAGGAGGTGGGGGACGCCTTGAGAGCCAAGTTTGAAGAAGGAAAAATCAAAAGAGAGGATATCTTCATAACGTCCAAGTTATGGTGCACGTTCCACCGCCCTGATCTCGTGGAGGAAGCTATCAAGACCTCGTTAAAGAATCTGGGTTTGGAGTACTTGGATCTCTATCTGATACACTGGCCCCAGGCCTTCAAGGAGGGCGACGACCTGTTCCCGAAGGATTCAGAGGACAAGTTCATCCCGTCGGCGGTGGACTACGTGGACACGTGGGGGGCGCTCGAGGCGCTGGTGGAGAAGGGCCTCACCCGCACCATAGGACTCTCCAACTTCAACAGGCGCCAGATTGAGAGAGTACTGGAAGTGGCCAAGATCAAGCCGGCTGTCCTCCAGATCGAAGTGCACCCCTACCTGAACCAGGAGAAGCTGTTCGAGTTCTGCAAGAGTCGCGACATCGCGGTGACGGCCTACTCCCCGCTGGGCTCCCCCGACCGCCCCGGCGCCGCCCCCACCGCCCCCCAGCTACTACAGGACCCCAGGCTACTCGCTCTCGCAGACAAACACTCCAAGACCACTGCGCAGATCCTGATCAGATACGCCATCGATCGCGGCATGATCGTGATCCCCAAGTCCGTGACGAGGAGCAGGATACAAGAGAACTTCAACGTGTTCGATTTCCAGCTGACCCTCGAGGACATCAAGCAGATATCTGCTCTGGAATGTAACGGGAGACTCTGCACTATGGGAGACACGCACCACCCCGACTACCCCTTCCACGACGAGTACTGA
Protein
MAAPVPVIFNNGRTCPVIGLGTWKSKPGEVTQAVKDAIDIGYRHVDCAHIYLNEKEVGDALRAKFEEGKIKREDIFITSKLWCTFHRPDLVEEAIKTSLKNLGLEYLDLYLIHWPQAFKEGDDLFPKDSEDKFIPSAVDYVDTWGALEALVEKGLTRTIGLSNFNRRQIERVLEVAKIKPAVLQIEVHPYLNQEKLFEFCKSRDIAVTAYSPLGSPDRPGAAPTAPQLLQDPRLLALADKHSKTTAQILIRYAIDRGMIVIPKSVTRSRIQENFNVFDFQLTLEDIKQISALECNGRLCTMGDTHHPDYPFHDEY
Summary
Uniprot
E5EVW4
U5KCL7
A0A291P0X7
A0A2W1B8U6
A0A2H1VL45
A0A2W1BCM3
+ More
A0A0N1PFE5 A0A1L8D6N9 A0A1L8D6W2 A0A076FRL7 D6WBY0 A0A0L7LJ80 A0A023EQM7 D6WBY2 Q17DM4 A0A182GCC4 A0A023EPH2 J3JX35 A0A088AGI8 A0A0T6AZE7 Q17DM5 U5KC00 A0A023EQ72 A0A1Q3FN84 A0A2A3EK43 V9ILR6 N6UG67 A0A1W4X5Y5 A0A182K591 A0A0N0BKC4 T1P8Q8 A0A1W4WL13 A0A182VI74 A0A182WUG5 A0A182KKP7 Q7PF06 A0A182HW13 A0A067QJF4 W5JBU8 A0A182JHG0 A0A1I9WLL7 A0A182IUH6 A0A1W4W9V6 T1DFA3 A0A194Q2M7 A0A182PHB5 A0A182FVL2 A0A2M4BTH9 A0A291P100 T1P7I4 A0A0L0BNI7 A0A2M3ZFC7 A0A2M4BTI2 A0A2M4CP84 A0A2A4J3U1 A0A2M4CPN6 A0A1I8NAC0 A0A151J3M8 A0A182YBI5 A0A2A4JAV0 A0A291P114 A0A182QI37 A0A0U4B4X4 A0A084WAD5 A0A2M4AF69 A0A182RUK3 A0A2W1BKW5 A0A182LYX7 U5EWM2 C4WSB5 A0A291P0V8 U5EWM3 A0A026WTR5 V5GQ24 F5BYH7 A0A2H8TF81 A0A0K8TNU1 D6WBY1 A0A291P0S6 A0A2H1W0K6 A0A2H1VAT7 J3JYE0 A0A158NZF8 A0A182WDU3 E2AB23 A0A151IW05 A0A1Y1N6N7 A0A151WRD3 F4WLB5 A0A2S2NGV3 Q17DM8 A0A195BMW2 A0A1B0A6Q4 J9K1Q6 A0A1D2MB39 Q563C9 A0A1A9W751 A0A195FYJ2 A0A182N3S8 A0A1I8P2H5
A0A0N1PFE5 A0A1L8D6N9 A0A1L8D6W2 A0A076FRL7 D6WBY0 A0A0L7LJ80 A0A023EQM7 D6WBY2 Q17DM4 A0A182GCC4 A0A023EPH2 J3JX35 A0A088AGI8 A0A0T6AZE7 Q17DM5 U5KC00 A0A023EQ72 A0A1Q3FN84 A0A2A3EK43 V9ILR6 N6UG67 A0A1W4X5Y5 A0A182K591 A0A0N0BKC4 T1P8Q8 A0A1W4WL13 A0A182VI74 A0A182WUG5 A0A182KKP7 Q7PF06 A0A182HW13 A0A067QJF4 W5JBU8 A0A182JHG0 A0A1I9WLL7 A0A182IUH6 A0A1W4W9V6 T1DFA3 A0A194Q2M7 A0A182PHB5 A0A182FVL2 A0A2M4BTH9 A0A291P100 T1P7I4 A0A0L0BNI7 A0A2M3ZFC7 A0A2M4BTI2 A0A2M4CP84 A0A2A4J3U1 A0A2M4CPN6 A0A1I8NAC0 A0A151J3M8 A0A182YBI5 A0A2A4JAV0 A0A291P114 A0A182QI37 A0A0U4B4X4 A0A084WAD5 A0A2M4AF69 A0A182RUK3 A0A2W1BKW5 A0A182LYX7 U5EWM2 C4WSB5 A0A291P0V8 U5EWM3 A0A026WTR5 V5GQ24 F5BYH7 A0A2H8TF81 A0A0K8TNU1 D6WBY1 A0A291P0S6 A0A2H1W0K6 A0A2H1VAT7 J3JYE0 A0A158NZF8 A0A182WDU3 E2AB23 A0A151IW05 A0A1Y1N6N7 A0A151WRD3 F4WLB5 A0A2S2NGV3 Q17DM8 A0A195BMW2 A0A1B0A6Q4 J9K1Q6 A0A1D2MB39 Q563C9 A0A1A9W751 A0A195FYJ2 A0A182N3S8 A0A1I8P2H5
Pubmed
19121390
21457781
24053512
28986331
28756777
26354079
+ More
24817326 18362917 19820115 26227816 24945155 17510324 26483478 22516182 23537049 20966253 12364791 14747013 17210077 24845553 20920257 23761445 27538518 26108605 25315136 25244985 26694822 24438588 24508170 21452889 26369729 21347285 20798317 28004739 21719571 27289101 15804580
24817326 18362917 19820115 26227816 24945155 17510324 26483478 22516182 23537049 20966253 12364791 14747013 17210077 24845553 20920257 23761445 27538518 26108605 25315136 25244985 26694822 24438588 24508170 21452889 26369729 21347285 20798317 28004739 21719571 27289101 15804580
EMBL
BABH01042122
HM012808
ADQ89807.1
KC007347
AGQ45612.1
MF687610
+ More
ATJ44536.1 KZ150224 PZC72072.1 ODYU01002976 SOQ41152.1 KZ150472 PZC70756.1 KQ458863 KPJ04704.1 GEYN01000018 JAV02111.1 GEYN01000022 JAV02107.1 KF960773 AII21975.1 KQ971311 EEZ98895.1 JTDY01000976 KOB75236.1 GAPW01002694 JAC10904.1 EEZ99011.1 CH477292 EAT44551.1 EAT44552.1 JXUM01009068 KQ560275 KXJ83379.1 GAPW01002693 JAC10905.1 BT127803 AEE62765.1 LJIG01022464 KRT80442.1 EAT44550.1 EAT44553.1 EAT44554.1 KC007349 AGQ45614.1 GAPW01002899 JAC10699.1 GFDL01006014 JAV29031.1 KZ288235 PBC31401.1 JR050380 AEY61259.1 APGK01021961 APGK01021962 KB740392 ENN80755.1 KQ435706 KOX80214.1 KA644505 AFP59134.1 AAAB01008804 EAA45511.3 APCN01001457 KK853419 KDR07716.1 ADMH02001680 ETN61456.1 KU932401 APA34037.1 AXCP01008792 GAMD01003201 JAA98389.1 KQ459580 KPI99254.1 GGFJ01007171 MBW56312.1 MF687613 ATJ44539.1 KA644506 AFP59135.1 JRES01001600 KNC21576.1 GGFM01006505 MBW27256.1 GGFJ01007172 MBW56313.1 GGFL01002974 MBW67152.1 NWSH01003180 PCG66817.1 GGFL01002973 MBW67151.1 KQ980275 KYN17034.1 NWSH01002318 PCG68542.1 MF687614 ATJ44540.1 AXCN02001099 KU218649 ALX00029.1 ATLV01022099 KE525328 KFB47179.1 GGFK01006115 MBW39436.1 KZ150102 PZC73496.1 AXCM01001195 GANO01001354 JAB58517.1 ABLF02027388 AK340122 BAH70785.1 MF687573 ATJ44499.1 GANO01001351 JAB58520.1 KK107109 EZA59353.1 GALX01004754 JAB63712.1 JF417982 AEB26313.1 GFXV01000143 MBW11948.1 GDAI01001569 JAI16034.1 EEZ99010.1 MF687581 ATJ44507.1 ODYU01005610 SOQ46635.1 ODYU01001555 SOQ37937.1 BT128266 AEE63226.1 ADTU01004584 ADTU01004585 GL438234 EFN69299.1 KQ980881 KYN11974.1 GEZM01013786 GEZM01013785 GEZM01013784 JAV92325.1 KQ982807 KYQ50361.1 GL888207 EGI65078.1 GGMR01003814 MBY16433.1 EAT44547.1 KQ976433 KYM87328.1 ABLF02027389 LJIJ01002106 ODM90218.1 AY947547 AAX85203.1 KQ981169 KYN45372.1
ATJ44536.1 KZ150224 PZC72072.1 ODYU01002976 SOQ41152.1 KZ150472 PZC70756.1 KQ458863 KPJ04704.1 GEYN01000018 JAV02111.1 GEYN01000022 JAV02107.1 KF960773 AII21975.1 KQ971311 EEZ98895.1 JTDY01000976 KOB75236.1 GAPW01002694 JAC10904.1 EEZ99011.1 CH477292 EAT44551.1 EAT44552.1 JXUM01009068 KQ560275 KXJ83379.1 GAPW01002693 JAC10905.1 BT127803 AEE62765.1 LJIG01022464 KRT80442.1 EAT44550.1 EAT44553.1 EAT44554.1 KC007349 AGQ45614.1 GAPW01002899 JAC10699.1 GFDL01006014 JAV29031.1 KZ288235 PBC31401.1 JR050380 AEY61259.1 APGK01021961 APGK01021962 KB740392 ENN80755.1 KQ435706 KOX80214.1 KA644505 AFP59134.1 AAAB01008804 EAA45511.3 APCN01001457 KK853419 KDR07716.1 ADMH02001680 ETN61456.1 KU932401 APA34037.1 AXCP01008792 GAMD01003201 JAA98389.1 KQ459580 KPI99254.1 GGFJ01007171 MBW56312.1 MF687613 ATJ44539.1 KA644506 AFP59135.1 JRES01001600 KNC21576.1 GGFM01006505 MBW27256.1 GGFJ01007172 MBW56313.1 GGFL01002974 MBW67152.1 NWSH01003180 PCG66817.1 GGFL01002973 MBW67151.1 KQ980275 KYN17034.1 NWSH01002318 PCG68542.1 MF687614 ATJ44540.1 AXCN02001099 KU218649 ALX00029.1 ATLV01022099 KE525328 KFB47179.1 GGFK01006115 MBW39436.1 KZ150102 PZC73496.1 AXCM01001195 GANO01001354 JAB58517.1 ABLF02027388 AK340122 BAH70785.1 MF687573 ATJ44499.1 GANO01001351 JAB58520.1 KK107109 EZA59353.1 GALX01004754 JAB63712.1 JF417982 AEB26313.1 GFXV01000143 MBW11948.1 GDAI01001569 JAI16034.1 EEZ99010.1 MF687581 ATJ44507.1 ODYU01005610 SOQ46635.1 ODYU01001555 SOQ37937.1 BT128266 AEE63226.1 ADTU01004584 ADTU01004585 GL438234 EFN69299.1 KQ980881 KYN11974.1 GEZM01013786 GEZM01013785 GEZM01013784 JAV92325.1 KQ982807 KYQ50361.1 GL888207 EGI65078.1 GGMR01003814 MBY16433.1 EAT44547.1 KQ976433 KYM87328.1 ABLF02027389 LJIJ01002106 ODM90218.1 AY947547 AAX85203.1 KQ981169 KYN45372.1
Proteomes
UP000005204
UP000053268
UP000007266
UP000037510
UP000008820
UP000069940
+ More
UP000249989 UP000005203 UP000242457 UP000019118 UP000192223 UP000075881 UP000053105 UP000075903 UP000076407 UP000075882 UP000007062 UP000075840 UP000027135 UP000000673 UP000075880 UP000075885 UP000069272 UP000037069 UP000218220 UP000095301 UP000078492 UP000076408 UP000075886 UP000030765 UP000075900 UP000075883 UP000007819 UP000053097 UP000005205 UP000075920 UP000000311 UP000075809 UP000007755 UP000078540 UP000092445 UP000094527 UP000091820 UP000078541 UP000075884 UP000095300
UP000249989 UP000005203 UP000242457 UP000019118 UP000192223 UP000075881 UP000053105 UP000075903 UP000076407 UP000075882 UP000007062 UP000075840 UP000027135 UP000000673 UP000075880 UP000075885 UP000069272 UP000037069 UP000218220 UP000095301 UP000078492 UP000076408 UP000075886 UP000030765 UP000075900 UP000075883 UP000007819 UP000053097 UP000005205 UP000075920 UP000000311 UP000075809 UP000007755 UP000078540 UP000092445 UP000094527 UP000091820 UP000078541 UP000075884 UP000095300
PRIDE
Pfam
PF00248 Aldo_ket_red
Interpro
SUPFAM
SSF51430
SSF51430
Gene 3D
ProteinModelPortal
E5EVW4
U5KCL7
A0A291P0X7
A0A2W1B8U6
A0A2H1VL45
A0A2W1BCM3
+ More
A0A0N1PFE5 A0A1L8D6N9 A0A1L8D6W2 A0A076FRL7 D6WBY0 A0A0L7LJ80 A0A023EQM7 D6WBY2 Q17DM4 A0A182GCC4 A0A023EPH2 J3JX35 A0A088AGI8 A0A0T6AZE7 Q17DM5 U5KC00 A0A023EQ72 A0A1Q3FN84 A0A2A3EK43 V9ILR6 N6UG67 A0A1W4X5Y5 A0A182K591 A0A0N0BKC4 T1P8Q8 A0A1W4WL13 A0A182VI74 A0A182WUG5 A0A182KKP7 Q7PF06 A0A182HW13 A0A067QJF4 W5JBU8 A0A182JHG0 A0A1I9WLL7 A0A182IUH6 A0A1W4W9V6 T1DFA3 A0A194Q2M7 A0A182PHB5 A0A182FVL2 A0A2M4BTH9 A0A291P100 T1P7I4 A0A0L0BNI7 A0A2M3ZFC7 A0A2M4BTI2 A0A2M4CP84 A0A2A4J3U1 A0A2M4CPN6 A0A1I8NAC0 A0A151J3M8 A0A182YBI5 A0A2A4JAV0 A0A291P114 A0A182QI37 A0A0U4B4X4 A0A084WAD5 A0A2M4AF69 A0A182RUK3 A0A2W1BKW5 A0A182LYX7 U5EWM2 C4WSB5 A0A291P0V8 U5EWM3 A0A026WTR5 V5GQ24 F5BYH7 A0A2H8TF81 A0A0K8TNU1 D6WBY1 A0A291P0S6 A0A2H1W0K6 A0A2H1VAT7 J3JYE0 A0A158NZF8 A0A182WDU3 E2AB23 A0A151IW05 A0A1Y1N6N7 A0A151WRD3 F4WLB5 A0A2S2NGV3 Q17DM8 A0A195BMW2 A0A1B0A6Q4 J9K1Q6 A0A1D2MB39 Q563C9 A0A1A9W751 A0A195FYJ2 A0A182N3S8 A0A1I8P2H5
A0A0N1PFE5 A0A1L8D6N9 A0A1L8D6W2 A0A076FRL7 D6WBY0 A0A0L7LJ80 A0A023EQM7 D6WBY2 Q17DM4 A0A182GCC4 A0A023EPH2 J3JX35 A0A088AGI8 A0A0T6AZE7 Q17DM5 U5KC00 A0A023EQ72 A0A1Q3FN84 A0A2A3EK43 V9ILR6 N6UG67 A0A1W4X5Y5 A0A182K591 A0A0N0BKC4 T1P8Q8 A0A1W4WL13 A0A182VI74 A0A182WUG5 A0A182KKP7 Q7PF06 A0A182HW13 A0A067QJF4 W5JBU8 A0A182JHG0 A0A1I9WLL7 A0A182IUH6 A0A1W4W9V6 T1DFA3 A0A194Q2M7 A0A182PHB5 A0A182FVL2 A0A2M4BTH9 A0A291P100 T1P7I4 A0A0L0BNI7 A0A2M3ZFC7 A0A2M4BTI2 A0A2M4CP84 A0A2A4J3U1 A0A2M4CPN6 A0A1I8NAC0 A0A151J3M8 A0A182YBI5 A0A2A4JAV0 A0A291P114 A0A182QI37 A0A0U4B4X4 A0A084WAD5 A0A2M4AF69 A0A182RUK3 A0A2W1BKW5 A0A182LYX7 U5EWM2 C4WSB5 A0A291P0V8 U5EWM3 A0A026WTR5 V5GQ24 F5BYH7 A0A2H8TF81 A0A0K8TNU1 D6WBY1 A0A291P0S6 A0A2H1W0K6 A0A2H1VAT7 J3JYE0 A0A158NZF8 A0A182WDU3 E2AB23 A0A151IW05 A0A1Y1N6N7 A0A151WRD3 F4WLB5 A0A2S2NGV3 Q17DM8 A0A195BMW2 A0A1B0A6Q4 J9K1Q6 A0A1D2MB39 Q563C9 A0A1A9W751 A0A195FYJ2 A0A182N3S8 A0A1I8P2H5
PDB
2PDW
E-value=4.38403e-99,
Score=921
Ontologies
PATHWAY
00040
Pentose and glucuronate interconversions - Bombyx mori (domestic silkworm)
00051 Fructose and mannose metabolism - Bombyx mori (domestic silkworm)
00052 Galactose metabolism - Bombyx mori (domestic silkworm)
00561 Glycerolipid metabolism - Bombyx mori (domestic silkworm)
00790 Folate biosynthesis - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
00051 Fructose and mannose metabolism - Bombyx mori (domestic silkworm)
00052 Galactose metabolism - Bombyx mori (domestic silkworm)
00561 Glycerolipid metabolism - Bombyx mori (domestic silkworm)
00790 Folate biosynthesis - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
GO
PANTHER
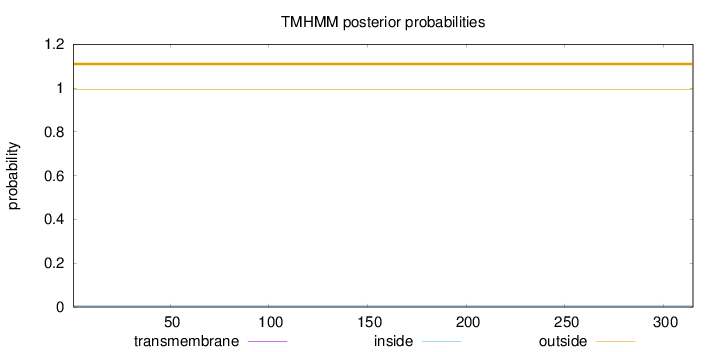
Topology
Length:
315
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.000780000000000001
Exp number, first 60 AAs:
0.00056
Total prob of N-in:
0.00600
outside
1 - 315
Population Genetic Test Statistics
Pi
271.079501
Theta
170.469829
Tajima's D
1.918424
CLR
0.035395
CSRT
0.869556522173891
Interpretation
Uncertain
