Gene
KWMTBOMO14893
Pre Gene Modal
BGIBMGA012178
Annotation
PREDICTED:_suppressor_of_fused_homolog_[Papilio_machaon]
Full name
Suppressor of fused homolog
Location in the cell
Nuclear Reliability : 1.139
Sequence
CDS
ATGTCCTGTCCGAGTGGCCCAAGGGCTCCTCATTCGTCGGGGGTGCCTTGCAACAGTGCCCTGTCATTCGTTGCGTCTGCGCAGCGAGTTCATGGACCGGAAAATGACGCTTGCAAACCTCAAGATGCCAAACAATCTGAACAACTAGTACCAGTAGCTCCGGCTGGCTTGAAAGCATTATGTGATGCTTGTTCCCAGCTGTACCCAGACCAGCCTAACCCTCTGCAGGTCACGACCAGGCTCAAATATTGGTTGGGAGGACAAGATCCTTTAGACTACATCAGCATGTACCGGAACCCAGGGAAACCGGAGGAAAATATACCACCACATTGGCACTACATCAGTTTCGGTCTGTCAGACCTTCACGGGGATGGGAGGGTCCACCCTCCGCCGGATCGCGCGGCGCTGTCCGCGGGCGCGCCCCTGCCGTCTGGCTGCGGCATCGAGCTGACGTTCCGGCTCGCGGCTGACGGGGCAGAGCACCCCCCGCTGTGGCCGGCGGCTCTCTTGCAGGCCTTGGCCAGATACGTGTTCACTACAGGTAACAAGCTGTGCGCCGGGGACCACATATCGTGGCATCGTCCGCTGGACGGCGAGAGGTCCGGGGGCCGCATGCGACACCTGCTGGTCGCGACAGAGCCGCAGCTGCGACACGCTCGCACGCCGCACGGCATCGTCACCTTCCTACAGATGGTGGGCTGCACCAGCAGAGAACTGAGGGCGGCCCAGAGGAGCTCCGTCTGCGAGGTACTGAAACTCATAGCGCAGGACCCGAGGTGCGGCGGGTGCTGGCTGGTGACGCGCATGGAGCGGCGGGTGTCGGTGCGGGCGGGGGGCGGCGCGGGGGGCGCGCTGGCGCAGCTGGCCGGCGTCTCCTGCGACGTGCGCTGGGCCGCGTGGGAGGATTCGACTGGTGAGTCGGTGGGCCGGGAGGATGCACAGGGGCTCGCCCACCTGGAGACCACGGTCACCGGACACATGTCCACGGACAGCTTCGGGATGTCGAGCATGGGAGAGGCGCTGGCGAAGCTCTCTGCCGAGGACCTCCCTGGGGATCCGGTGCAGTACCTGAGCGGGGTCCACGTCGTCCTGAGCCCGGAAGGCGCCGCCCTGCTGCCCCTGGCCGTCGAGGGCCGCATTCTACACGGAGCGCACTTCACGTGGCGCGGCGAGTCGGCGGCCGTCACGCTGGTGGCGCCCGGCGTGCACGGCTGCTTCGTGAGCACCGACGCGCCGCGCGCCGCCCGGGGCTCCTGGCTGCAGATATTAGTGAACAAGAGCCTCGCCTCGGATATACTGGCCCAGCGGAGCGCCCTCTGTGACGACGCGCACTCCGACAGCGAGGAGGACGCCCCCGCCCCCGCCCCCGCGCCCCCCCGCACCGTGCGCTGGCCCGAGCACGGGCTGGCCATCACCGTGCGCGCCTGA
Protein
MSCPSGPRAPHSSGVPCNSALSFVASAQRVHGPENDACKPQDAKQSEQLVPVAPAGLKALCDACSQLYPDQPNPLQVTTRLKYWLGGQDPLDYISMYRNPGKPEENIPPHWHYISFGLSDLHGDGRVHPPPDRAALSAGAPLPSGCGIELTFRLAADGAEHPPLWPAALLQALARYVFTTGNKLCAGDHISWHRPLDGERSGGRMRHLLVATEPQLRHARTPHGIVTFLQMVGCTSRELRAAQRSSVCEVLKLIAQDPRCGGCWLVTRMERRVSVRAGGGAGGALAQLAGVSCDVRWAAWEDSTGESVGREDAQGLAHLETTVTGHMSTDSFGMSSMGEALAKLSAEDLPGDPVQYLSGVHVVLSPEGAALLPLAVEGRILHGAHFTWRGESAAVTLVAPGVHGCFVSTDAPRAARGSWLQILVNKSLASDILAQRSALCDDAHSDSEEDAPAPAPAPPRTVRWPEHGLAITVRA
Summary
Description
Negative regulator in the hedgehog/smoothened signaling pathway (PubMed:16155214, PubMed:16459298). Down-regulates GLI1-mediated transactivation of target genes (PubMed:11960000). Part of a corepressor complex that acts on DNA-bound GLI1 (PubMed:11960000). May also act by linking GLI1 to BTRC and thereby targeting GLI1 to degradation by the proteasome (By similarity). Sequesters GLI1, GLI2 and GLI3 in the cytoplasm, this effect is overcome by binding of STK36 to both SUFU and a GLI protein (PubMed:10531011, PubMed:16459298). Negative regulator of beta-catenin signaling (PubMed:11477086). Regulates the formation of either the repressor form (GLI3R) or the activator form (GLI3A) of the full-length form of GLI3 (GLI3FL) (PubMed:10531011, PubMed:20360384). GLI3FL is complexed with SUFU in the cytoplasm and is maintained in a neutral state (PubMed:10531011, PubMed:20360384). Without the Hh signal, the SUFU-GLI3 complex is recruited to cilia, leading to the efficient processing of GLI3FL into GLI3R (PubMed:10531011, PubMed:20360384). When Hh signaling is initiated, SUFU dissociates from GLI3FL and the latter translocates to the nucleus, where it is phosphorylated, destabilized, and converted to a transcriptional activator (GLI3A) (PubMed:10531011, PubMed:20360384). Required for normal embryonic development (PubMed:16155214, PubMed:16459298). Required for the proper formation of hair follicles and the control of epidermal differentiation (PubMed:16155214, PubMed:16459298, PubMed:23034632).
Negative regulator in the hedgehog signaling pathway. Down-regulates GLI1-mediated transactivation of target genes. Part of a corepressor complex that acts on DNA-bound GLI1. May also act by linking GLI1 to BTRC and thereby targeting GLI1 to degradation by the proteasome. Sequesters GLI1, GLI2 and GLI3 in the cytoplasm, this effect is overcome by binding of STK36 to both SUFU and a GLI protein. Negative regulator of beta-catenin signaling. Regulates the formation of either the repressor form (GLI3R) or the activator form (GLI3A) of the full-length form of GLI3 (GLI3FL). GLI3FL is complexed with SUFU in the cytoplasm and is maintained in a neutral state. Without the Hh signal, the SUFU-GLI3 complex is recruited to cilia, leading to the efficient processing of GLI3FL into GLI3R. When Hh signaling is initiated, SUFU dissociates from GLI3FL and the latter translocates to the nucleus, where it is phosphorylated, destabilized, and converted to a transcriptional activator (GLI3A).
Negative regulator in the hedgehog signaling pathway. Down-regulates GLI1-mediated transactivation of target genes. Part of a corepressor complex that acts on DNA-bound GLI1. May also act by linking GLI1 to BTRC and thereby targeting GLI1 to degradation by the proteasome. Sequesters GLI1, GLI2 and GLI3 in the cytoplasm, this effect is overcome by binding of STK36 to both SUFU and a GLI protein. Negative regulator of beta-catenin signaling. Regulates the formation of either the repressor form (GLI3R) or the activator form (GLI3A) of the full-length form of GLI3 (GLI3FL). GLI3FL is complexed with SUFU in the cytoplasm and is maintained in a neutral state. Without the Hh signal, the SUFU-GLI3 complex is recruited to cilia, leading to the efficient processing of GLI3FL into GLI3R. When Hh signaling is initiated, SUFU dissociates from GLI3FL and the latter translocates to the nucleus, where it is phosphorylated, destabilized, and converted to a transcriptional activator (GLI3A).
Subunit
May form homodimers (By similarity). Interacts with ULK3; inactivating the protein kinase activity of ULK3. Interacts with RAB23 (By similarity). Part of a DNA-bound corepressor complex containing SAP18, GLI1 and SIN3 (PubMed:11960000). Part of a complex containing CTNNB1 (PubMed:11477086). Binds BTRC, GLI2, GLI3, SAP18 and STK36 (PubMed:20360384, PubMed:23034632). Binds both free and DNA-bound GLI1 (PubMed:10531011). Interacts with KIF7 (PubMed:19592253). Interacts with GLI3FL and this interaction regulates the formation of either repressor or activator forms of GLI3 (PubMed:20360384). Its association with GLI3FL is regulated by Hh signaling and dissociation of the SUFU-GLI3 interaction requires the presence of the ciliary motor KIF3A (PubMed:20360384).
Similarity
Belongs to the SUFU family.
Keywords
Acetylation
Alternative splicing
Complete proteome
Cytoplasm
Developmental protein
Isopeptide bond
Nucleus
Phosphoprotein
Reference proteome
Ubl conjugation
Feature
chain Suppressor of fused homolog
splice variant In isoform 2.
splice variant In isoform 2.
Uniprot
H9JRL8
A0A2H1V7Q8
S4NSH3
A0A0N1IA38
A0A212EJU1
A0A0N1PG72
+ More
B0W2C6 A0A1W4XHY0 A0A1Y1N1A7 A0A182PQV3 A0A084VS45 V4AT21 A0A0P4W721 A0A182GH08 Q17FL2 A0A2S2R1B4 A0A182I0B1 A0A182RXK3 A0A182VR28 Q7PYI0 A0A182KLZ3 A0A182U4W4 A0A182XBH1 A0A182JR76 A0A182VAV4 A0A023F1Y6 A0A182MI26 A0A2G8L5D3 D1LXF3 T1I0U2 A0A0V0GCH3 A0A3R7M5B9 A0A224XQB4 A0A182J8K8 A0A182N4B4 A0A182QI59 A0A1W2WEY3 V8PFR0 A0A2C9JDF8 A0A0A9YJX6 A0A146LDP5 A0A0K8TGC4 A0A2M4CZF7 A0A139WMW0 H2YPK1 A0A3B4VMG5 A0A3Q3L3C0 F6Z6K5 A0A3Q3KSD7 A0A3Q3L3Q4 A0A210Q2A3 A0A3B3S7Y6 W4XDB0 E9FSV5 A0A1X7VXQ0 A0A2M4AX36 A0A0P4VQE8 U3K235 A0A2L2YFU1 Q9Z0P7-4 Q3U0Z8 A0A2U4BQG9 Q9Z0P7 Q4V8C6 A0A2Y9N5Y6 A0A093PQE9 A0A2Y9FMI7 A0A384AJ11 A0A1S3WKV4 A0A340YDG8 G3SVH6 A0A3Q0CYP9 A0A3Q7RT42 A0A3Q7W554 G3R6K6 A0A2U4BQN1 A0A2K6CAP0 A0A2K5WNJ5 A0A096P6F4 A0A2K5N8H0 H2Q2H8 A0A2K6NDB1 A0A2K6TU03 A0A1S3F8U5 A0A1S3A7Y4 G1RYV3 U3EN30 A0A2Y9MRM2 A0A2Y9J8H7 A0A340YBN2 A0A2Y9FN18 A0A384AIX4 G7N110
B0W2C6 A0A1W4XHY0 A0A1Y1N1A7 A0A182PQV3 A0A084VS45 V4AT21 A0A0P4W721 A0A182GH08 Q17FL2 A0A2S2R1B4 A0A182I0B1 A0A182RXK3 A0A182VR28 Q7PYI0 A0A182KLZ3 A0A182U4W4 A0A182XBH1 A0A182JR76 A0A182VAV4 A0A023F1Y6 A0A182MI26 A0A2G8L5D3 D1LXF3 T1I0U2 A0A0V0GCH3 A0A3R7M5B9 A0A224XQB4 A0A182J8K8 A0A182N4B4 A0A182QI59 A0A1W2WEY3 V8PFR0 A0A2C9JDF8 A0A0A9YJX6 A0A146LDP5 A0A0K8TGC4 A0A2M4CZF7 A0A139WMW0 H2YPK1 A0A3B4VMG5 A0A3Q3L3C0 F6Z6K5 A0A3Q3KSD7 A0A3Q3L3Q4 A0A210Q2A3 A0A3B3S7Y6 W4XDB0 E9FSV5 A0A1X7VXQ0 A0A2M4AX36 A0A0P4VQE8 U3K235 A0A2L2YFU1 Q9Z0P7-4 Q3U0Z8 A0A2U4BQG9 Q9Z0P7 Q4V8C6 A0A2Y9N5Y6 A0A093PQE9 A0A2Y9FMI7 A0A384AJ11 A0A1S3WKV4 A0A340YDG8 G3SVH6 A0A3Q0CYP9 A0A3Q7RT42 A0A3Q7W554 G3R6K6 A0A2U4BQN1 A0A2K6CAP0 A0A2K5WNJ5 A0A096P6F4 A0A2K5N8H0 H2Q2H8 A0A2K6NDB1 A0A2K6TU03 A0A1S3F8U5 A0A1S3A7Y4 G1RYV3 U3EN30 A0A2Y9MRM2 A0A2Y9J8H7 A0A340YBN2 A0A2Y9FN18 A0A384AIX4 G7N110
Pubmed
19121390
23622113
26354079
22118469
28004739
24438588
+ More
23254933 26483478 17510324 12364791 14747013 17210077 20966253 25474469 29023486 24297900 15562597 25401762 26823975 18362917 19820115 12481130 15114417 28812685 29240929 21292972 27129103 26561354 10531011 11252182 11557033 16141072 15489334 11477086 11960000 16155214 16459298 19592253 21183079 20360384 23034632 10349636 11042159 11076861 11217851 12466851 12040188 16141073 15057822 15632090 28071753 22398555 16136131 25362486 25243066 22002653
23254933 26483478 17510324 12364791 14747013 17210077 20966253 25474469 29023486 24297900 15562597 25401762 26823975 18362917 19820115 12481130 15114417 28812685 29240929 21292972 27129103 26561354 10531011 11252182 11557033 16141072 15489334 11477086 11960000 16155214 16459298 19592253 21183079 20360384 23034632 10349636 11042159 11076861 11217851 12466851 12040188 16141073 15057822 15632090 28071753 22398555 16136131 25362486 25243066 22002653
EMBL
BABH01031732
BABH01031733
BABH01031734
ODYU01001096
SOQ36847.1
GAIX01014097
+ More
JAA78463.1 KQ459988 KPJ18793.1 AGBW02014393 OWR41757.1 KQ458863 KPJ04723.1 DS231826 EDS28741.1 GEZM01020189 JAV89327.1 ATLV01015809 KE525036 KFB40789.1 KB201362 ESO96856.1 GDRN01086756 JAI61123.1 JXUM01062750 KQ562214 KXJ76391.1 CH477270 EAT45331.1 GGMS01014573 MBY83776.1 APCN01002058 AAAB01008987 EAA01100.1 GBBI01003145 JAC15567.1 AXCM01008324 MRZV01000214 PIK55464.1 GU076130 ACY92659.1 ACPB03010575 GECL01000607 JAP05517.1 QCYY01002039 ROT73307.1 GFTR01006235 JAW10191.1 AXCN02000031 AZIM01000124 ETE73185.1 GBHO01017376 GBHO01013779 GBHO01013772 JAG26228.1 JAG29825.1 JAG29832.1 GDHC01013004 JAQ05625.1 GBRD01001183 JAG64638.1 GGFL01006433 MBW70611.1 KQ971312 KYB29272.1 EAAA01002614 NEDP02005207 OWF42883.1 AAGJ04030573 GL732524 EFX89751.1 GGFK01011837 MBW45158.1 GDKW01003205 JAI53390.1 AGTO01020393 LC379607 IAAA01021144 BBD75244.1 LAA06988.1 AF134893 AJ131692 AJ308625 AJ308626 AJ308627 AJ308628 AJ308629 AJ308630 AJ308632 AJ308634 AJ308635 AJ308633 AJ308631 AK015885 AK047603 BC048168 BC056997 AK156410 CH466534 BAE33703.1 EDL42000.1 AC097694 BC097447 CH473986 AAH97447.1 EDL94350.1 KL669486 KFW76470.1 CABD030075842 CABD030075843 CABD030075844 CABD030075845 CABD030075846 CABD030075847 CABD030075848 CABD030075849 AQIA01072379 AQIA01072380 AQIA01072381 AHZZ02035970 AACZ04051127 AACZ04051128 AACZ04051129 ADFV01163581 ADFV01163582 ADFV01163583 ADFV01163584 ADFV01163585 ADFV01163586 ADFV01163587 ADFV01163588 ADFV01163589 ADFV01163590 GAMR01010612 JAB23320.1 CM001261 EHH19335.1
JAA78463.1 KQ459988 KPJ18793.1 AGBW02014393 OWR41757.1 KQ458863 KPJ04723.1 DS231826 EDS28741.1 GEZM01020189 JAV89327.1 ATLV01015809 KE525036 KFB40789.1 KB201362 ESO96856.1 GDRN01086756 JAI61123.1 JXUM01062750 KQ562214 KXJ76391.1 CH477270 EAT45331.1 GGMS01014573 MBY83776.1 APCN01002058 AAAB01008987 EAA01100.1 GBBI01003145 JAC15567.1 AXCM01008324 MRZV01000214 PIK55464.1 GU076130 ACY92659.1 ACPB03010575 GECL01000607 JAP05517.1 QCYY01002039 ROT73307.1 GFTR01006235 JAW10191.1 AXCN02000031 AZIM01000124 ETE73185.1 GBHO01017376 GBHO01013779 GBHO01013772 JAG26228.1 JAG29825.1 JAG29832.1 GDHC01013004 JAQ05625.1 GBRD01001183 JAG64638.1 GGFL01006433 MBW70611.1 KQ971312 KYB29272.1 EAAA01002614 NEDP02005207 OWF42883.1 AAGJ04030573 GL732524 EFX89751.1 GGFK01011837 MBW45158.1 GDKW01003205 JAI53390.1 AGTO01020393 LC379607 IAAA01021144 BBD75244.1 LAA06988.1 AF134893 AJ131692 AJ308625 AJ308626 AJ308627 AJ308628 AJ308629 AJ308630 AJ308632 AJ308634 AJ308635 AJ308633 AJ308631 AK015885 AK047603 BC048168 BC056997 AK156410 CH466534 BAE33703.1 EDL42000.1 AC097694 BC097447 CH473986 AAH97447.1 EDL94350.1 KL669486 KFW76470.1 CABD030075842 CABD030075843 CABD030075844 CABD030075845 CABD030075846 CABD030075847 CABD030075848 CABD030075849 AQIA01072379 AQIA01072380 AQIA01072381 AHZZ02035970 AACZ04051127 AACZ04051128 AACZ04051129 ADFV01163581 ADFV01163582 ADFV01163583 ADFV01163584 ADFV01163585 ADFV01163586 ADFV01163587 ADFV01163588 ADFV01163589 ADFV01163590 GAMR01010612 JAB23320.1 CM001261 EHH19335.1
Proteomes
UP000005204
UP000053240
UP000007151
UP000053268
UP000002320
UP000192223
+ More
UP000075885 UP000030765 UP000030746 UP000069940 UP000249989 UP000008820 UP000075840 UP000075900 UP000075920 UP000007062 UP000075882 UP000075902 UP000076407 UP000075881 UP000075903 UP000075883 UP000230750 UP000015103 UP000283509 UP000075880 UP000075884 UP000075886 UP000076420 UP000007266 UP000007875 UP000261420 UP000261640 UP000008144 UP000242188 UP000261540 UP000007110 UP000000305 UP000007879 UP000016665 UP000000589 UP000245320 UP000002494 UP000248483 UP000053258 UP000261681 UP000079721 UP000265300 UP000007646 UP000189706 UP000286640 UP000286642 UP000001519 UP000233120 UP000233100 UP000028761 UP000233060 UP000002277 UP000233200 UP000233220 UP000081671 UP000001073 UP000248482
UP000075885 UP000030765 UP000030746 UP000069940 UP000249989 UP000008820 UP000075840 UP000075900 UP000075920 UP000007062 UP000075882 UP000075902 UP000076407 UP000075881 UP000075903 UP000075883 UP000230750 UP000015103 UP000283509 UP000075880 UP000075884 UP000075886 UP000076420 UP000007266 UP000007875 UP000261420 UP000261640 UP000008144 UP000242188 UP000261540 UP000007110 UP000000305 UP000007879 UP000016665 UP000000589 UP000245320 UP000002494 UP000248483 UP000053258 UP000261681 UP000079721 UP000265300 UP000007646 UP000189706 UP000286640 UP000286642 UP000001519 UP000233120 UP000233100 UP000028761 UP000233060 UP000002277 UP000233200 UP000233220 UP000081671 UP000001073 UP000248482
Interpro
SUPFAM
SSF103359
SSF103359
Gene 3D
ProteinModelPortal
H9JRL8
A0A2H1V7Q8
S4NSH3
A0A0N1IA38
A0A212EJU1
A0A0N1PG72
+ More
B0W2C6 A0A1W4XHY0 A0A1Y1N1A7 A0A182PQV3 A0A084VS45 V4AT21 A0A0P4W721 A0A182GH08 Q17FL2 A0A2S2R1B4 A0A182I0B1 A0A182RXK3 A0A182VR28 Q7PYI0 A0A182KLZ3 A0A182U4W4 A0A182XBH1 A0A182JR76 A0A182VAV4 A0A023F1Y6 A0A182MI26 A0A2G8L5D3 D1LXF3 T1I0U2 A0A0V0GCH3 A0A3R7M5B9 A0A224XQB4 A0A182J8K8 A0A182N4B4 A0A182QI59 A0A1W2WEY3 V8PFR0 A0A2C9JDF8 A0A0A9YJX6 A0A146LDP5 A0A0K8TGC4 A0A2M4CZF7 A0A139WMW0 H2YPK1 A0A3B4VMG5 A0A3Q3L3C0 F6Z6K5 A0A3Q3KSD7 A0A3Q3L3Q4 A0A210Q2A3 A0A3B3S7Y6 W4XDB0 E9FSV5 A0A1X7VXQ0 A0A2M4AX36 A0A0P4VQE8 U3K235 A0A2L2YFU1 Q9Z0P7-4 Q3U0Z8 A0A2U4BQG9 Q9Z0P7 Q4V8C6 A0A2Y9N5Y6 A0A093PQE9 A0A2Y9FMI7 A0A384AJ11 A0A1S3WKV4 A0A340YDG8 G3SVH6 A0A3Q0CYP9 A0A3Q7RT42 A0A3Q7W554 G3R6K6 A0A2U4BQN1 A0A2K6CAP0 A0A2K5WNJ5 A0A096P6F4 A0A2K5N8H0 H2Q2H8 A0A2K6NDB1 A0A2K6TU03 A0A1S3F8U5 A0A1S3A7Y4 G1RYV3 U3EN30 A0A2Y9MRM2 A0A2Y9J8H7 A0A340YBN2 A0A2Y9FN18 A0A384AIX4 G7N110
B0W2C6 A0A1W4XHY0 A0A1Y1N1A7 A0A182PQV3 A0A084VS45 V4AT21 A0A0P4W721 A0A182GH08 Q17FL2 A0A2S2R1B4 A0A182I0B1 A0A182RXK3 A0A182VR28 Q7PYI0 A0A182KLZ3 A0A182U4W4 A0A182XBH1 A0A182JR76 A0A182VAV4 A0A023F1Y6 A0A182MI26 A0A2G8L5D3 D1LXF3 T1I0U2 A0A0V0GCH3 A0A3R7M5B9 A0A224XQB4 A0A182J8K8 A0A182N4B4 A0A182QI59 A0A1W2WEY3 V8PFR0 A0A2C9JDF8 A0A0A9YJX6 A0A146LDP5 A0A0K8TGC4 A0A2M4CZF7 A0A139WMW0 H2YPK1 A0A3B4VMG5 A0A3Q3L3C0 F6Z6K5 A0A3Q3KSD7 A0A3Q3L3Q4 A0A210Q2A3 A0A3B3S7Y6 W4XDB0 E9FSV5 A0A1X7VXQ0 A0A2M4AX36 A0A0P4VQE8 U3K235 A0A2L2YFU1 Q9Z0P7-4 Q3U0Z8 A0A2U4BQG9 Q9Z0P7 Q4V8C6 A0A2Y9N5Y6 A0A093PQE9 A0A2Y9FMI7 A0A384AJ11 A0A1S3WKV4 A0A340YDG8 G3SVH6 A0A3Q0CYP9 A0A3Q7RT42 A0A3Q7W554 G3R6K6 A0A2U4BQN1 A0A2K6CAP0 A0A2K5WNJ5 A0A096P6F4 A0A2K5N8H0 H2Q2H8 A0A2K6NDB1 A0A2K6TU03 A0A1S3F8U5 A0A1S3A7Y4 G1RYV3 U3EN30 A0A2Y9MRM2 A0A2Y9J8H7 A0A340YBN2 A0A2Y9FN18 A0A384AIX4 G7N110
PDB
4BLA
E-value=7.58403e-77,
Score=731
Ontologies
GO
GO:0008134
GO:0005737
GO:0005634
GO:0043433
GO:0042994
GO:0016021
GO:0002088
GO:0030097
GO:0033333
GO:0007224
GO:0008270
GO:0035301
GO:0045879
GO:1901621
GO:0008013
GO:0019901
GO:0043588
GO:0035904
GO:0000122
GO:2000059
GO:0021775
GO:0005929
GO:0001843
GO:0060976
GO:0021776
GO:0001947
GO:0003281
GO:0005829
GO:0007368
GO:0021513
GO:0007275
GO:0005515
GO:0006457
GO:0051082
GO:0003676
GO:0016701
GO:0006355
GO:0003824
GO:0008152
GO:0016491
GO:0015930
PANTHER
Topology
Subcellular location
Cytoplasm
Nucleus
Nucleus
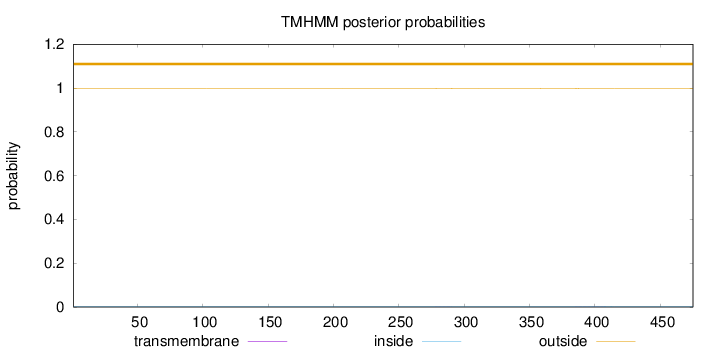
Length:
475
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01873
Exp number, first 60 AAs:
0.00019
Total prob of N-in:
0.00125
outside
1 - 475
Population Genetic Test Statistics
Pi
23.689637
Theta
168.162872
Tajima's D
0.117889
CLR
0.177558
CSRT
0.407629618519074
Interpretation
Uncertain
