Gene
KWMTBOMO14887 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA012182
Annotation
PREDICTED:_T-complex_protein_1_subunit_eta_isoform_X2_[Papilio_xuthus]
Full name
T-complex protein 1 subunit eta
Alternative Name
CCT-eta
Location in the cell
Cytoplasmic Reliability : 3.352
Sequence
CDS
ATGCAACCGCAAATACTTTTACTGAGAGAGGGAACGGACCAGACGCAGGGCAAGCCCCAGCTCGTCTCCAACATCAATGCCTGTCAACTTGTTGTCGATGCGGTCAGAACCACCCTGGGTCCCCGTGGTATGGACAAGCTGATTGTAGATCACAATGGAAAGGCGGTTATATCAAACGATGGTGCAACGATCATGAAGCTACTGGATATCATTCACCCTGCCGCTAAAACTCTTGTGGATATTGCAAAGTCTCAAGATGCCGAGGTTGGCGATGGAACCACCTCGGTAGTGATATTGGCCGGTGAAATTCTGAAGAGGTTGAAGCCGTTTGTTGAAGAAGGTGTTCACCCTCGTGTTCTGATCAGAGCAGTCAGAACAGCGTCAAGACTGGCCATTGAGAAAATCAAGGAGCAAGCTGTGAAAATTGATAATAAGTCGCCTGAGGAACAAAGAGATCTCCTATTGAAGTGTGCATCAACAGCGATGTCATCGAAACTGATTCATCAACAGAAGGATCACTTCTCTAAGATTGTTGTTGATGCTGTCCTGTCTTTGGACACTCCACTTCTGCCTCTCGACATGATTGGAATCAAGAAGGTTCCCGGTGGAGCTCTTGAAGATTCGTTCCTGGTTCCCGGAGTGGCGTTCAAGAAAACTTTCAGTTACGCTGGCTTTGAGATGCAGCCGAAGACATACAAAGACTGCAAGATCGCACTCCTCAATATTGAGCTGGAATTGAAAGCTGAAAGAGACAATGCCGAGGTTCGAGTAAACAACGTGAGGGAATATCAGAAGGTGGTGGATGCTGAGTGGCAGATCTTATACGAGAAGCTCGCCGCTTTGCATGCTAGCGGAGCAAATGTGGTCCTCAGCAAGCTGCCCATCGGAGATGTTGCCACACAATACTTTGCGGACCGAGATATGTTCTGCGCTGGACGAGTGCCCGAGGAAGACATGCGGCGTACGGCGCGCGCTTGCGGGGGAGCCGTACTCGCCTCAACGAGGGACCTCACGCCTCAAGTTCTAGGTCACACGGCGCTCTTCACGGAGAAACAGCTTGGAGCTGACAGATACAACTTCTTTACTGGCTGTCCGGGTGCCAAGACATGTACATTGATCCTGCGAGGTGGTGCGGAACAGTTCTTGGAGGAGACGGAGCGTTCCCTGCACGATGCCATCATGATAGTCCGGAGAACGATCAAGAATGATGCTGTAGTGGCTGGCGGAGGAGCAGTAGACATGCAAATCTCTGCCCATCTCCGGTCTCACTCTCTACAACTGGCTGGCAAGGAGCAGTTGGCTGTAGCGGCAGTGGCCCGAGCCTTTGAGGCCATCCCTCGTCAGCTGGCAGACAATGCAGGCCTGGACGGGACCGGACTCCTCAATAAACTAAGGCAGAAGCACGCCGCTGGGGAGCACACGCACGGTGTGGACGTTATCAGTGGAGAGGTGGTGGATAACTTTGCGAAGTGCGTGTGGGAGCCGGCCCTCGTCAAGCTTAACGCCGTGAGTGCCGCCTGCGAGGCCACCGCTCAGATTCTATCAGTGGATGAGACCATCAAGAATGTCAAGGGCGGTGAAGAGGCGCAGATGCCTGGTCGCGGCATGGGCCGCCCTCGTATGGGATAG
Protein
MQPQILLLREGTDQTQGKPQLVSNINACQLVVDAVRTTLGPRGMDKLIVDHNGKAVISNDGATIMKLLDIIHPAAKTLVDIAKSQDAEVGDGTTSVVILAGEILKRLKPFVEEGVHPRVLIRAVRTASRLAIEKIKEQAVKIDNKSPEEQRDLLLKCASTAMSSKLIHQQKDHFSKIVVDAVLSLDTPLLPLDMIGIKKVPGGALEDSFLVPGVAFKKTFSYAGFEMQPKTYKDCKIALLNIELELKAERDNAEVRVNNVREYQKVVDAEWQILYEKLAALHASGANVVLSKLPIGDVATQYFADRDMFCAGRVPEEDMRRTARACGGAVLASTRDLTPQVLGHTALFTEKQLGADRYNFFTGCPGAKTCTLILRGGAEQFLEETERSLHDAIMIVRRTIKNDAVVAGGGAVDMQISAHLRSHSLQLAGKEQLAVAAVARAFEAIPRQLADNAGLDGTGLLNKLRQKHAAGEHTHGVDVISGEVVDNFAKCVWEPALVKLNAVSAACEATAQILSVDETIKNVKGGEEAQMPGRGMGRPRMG
Summary
Description
Molecular chaperone; assists the folding of proteins upon ATP hydrolysis. Known to play a role, in vitro, in the folding of actin and tubulin.
Subunit
Heterooligomeric complex of about 850 to 900 kDa that forms two stacked rings, 12 to 16 nm in diameter.
Similarity
Belongs to the TCP-1 chaperonin family.
Uniprot
H9JRM2
A0A2A4KB20
A0A212EJW0
A0A0N1I5L4
A0A0L0C7B5
A0A1L8DUA7
+ More
U5EYW9 A0A1Y9H2B3 A0A0A1WZV3 A0A182F992 A0A2M3ZGM4 W8B7A3 B4KDM5 A0A182Y3V9 W5JAM7 A0A034VMZ5 A0A2M4A661 A0A2M4BJF5 T1DGV8 A0A182UQ34 T1PDA9 A0A182U284 A0A182XIU0 Q7PGM0 A0A182VZP9 A0A1B6LLS6 A0A182HTH9 A0A182QWQ4 A0A084VWM2 A0A0K8WKZ7 A0A182P504 A0A1L8ECI8 B4LWV7 A0A1B6F0B1 A0A182JW05 A0A1I8NV51 B3LXX7 A0A232F081 A0A1Q3F8H6 Q9VHL2 A0A182M116 A0A023EUT4 A0A1W4W2H7 B4NB31 A0A1B0CJ02 A0A336LHQ7 Q17P27 E0V947 A0A1B6CDI9 B4PTW7 B3P4R8 D3TLV7 A0A182J8C4 A0A0M3QYN8 A0A0A9Y855 A0A1W4X638 B0WFH0 A0A182RSY3 A0A1Y1K9S1 A0A1J1IYW5 A0A3B0JP67 D0QWN5 J3JWM9 Q294I9 A0A0P6IC77 A0A164VFC7 B4GL19 E9FX08 A0A023F4G7 A0A0V0G3V3 A0A0P6AN96 A0A069DV50 A0A0P5BTH5 A0A224XHV4 V5G6I1 A0A151J6G8 A0A0P4VTR7 A0A1S3I1C0 D6WBZ5 T1IDR0 A0A385L3J3 J9PNK3 V5RDK5 A0A195FJ69 A0A026W0P5 A0A0P4WFK2 A0A195C9J2 F4WX99 A0A151WMY4 A0A3L8DNN7 A0A158NU49 A0A2J7PBY2 A0A1B6ENH8 T1FNQ7 Q95V46 A0A0P5B6L9 E2B249 E9IQ54 A0A0J7L5X5 A0A293LGP1
U5EYW9 A0A1Y9H2B3 A0A0A1WZV3 A0A182F992 A0A2M3ZGM4 W8B7A3 B4KDM5 A0A182Y3V9 W5JAM7 A0A034VMZ5 A0A2M4A661 A0A2M4BJF5 T1DGV8 A0A182UQ34 T1PDA9 A0A182U284 A0A182XIU0 Q7PGM0 A0A182VZP9 A0A1B6LLS6 A0A182HTH9 A0A182QWQ4 A0A084VWM2 A0A0K8WKZ7 A0A182P504 A0A1L8ECI8 B4LWV7 A0A1B6F0B1 A0A182JW05 A0A1I8NV51 B3LXX7 A0A232F081 A0A1Q3F8H6 Q9VHL2 A0A182M116 A0A023EUT4 A0A1W4W2H7 B4NB31 A0A1B0CJ02 A0A336LHQ7 Q17P27 E0V947 A0A1B6CDI9 B4PTW7 B3P4R8 D3TLV7 A0A182J8C4 A0A0M3QYN8 A0A0A9Y855 A0A1W4X638 B0WFH0 A0A182RSY3 A0A1Y1K9S1 A0A1J1IYW5 A0A3B0JP67 D0QWN5 J3JWM9 Q294I9 A0A0P6IC77 A0A164VFC7 B4GL19 E9FX08 A0A023F4G7 A0A0V0G3V3 A0A0P6AN96 A0A069DV50 A0A0P5BTH5 A0A224XHV4 V5G6I1 A0A151J6G8 A0A0P4VTR7 A0A1S3I1C0 D6WBZ5 T1IDR0 A0A385L3J3 J9PNK3 V5RDK5 A0A195FJ69 A0A026W0P5 A0A0P4WFK2 A0A195C9J2 F4WX99 A0A151WMY4 A0A3L8DNN7 A0A158NU49 A0A2J7PBY2 A0A1B6ENH8 T1FNQ7 Q95V46 A0A0P5B6L9 E2B249 E9IQ54 A0A0J7L5X5 A0A293LGP1
Pubmed
19121390
22118469
26354079
26108605
25830018
24495485
+ More
17994087 25244985 20920257 23761445 25348373 12364791 14747013 17210077 24438588 28648823 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24945155 26483478 17510324 20566863 17550304 20353571 25401762 26823975 28004739 19859648 22516182 23537049 15632085 21292972 25474469 26334808 27129103 18362917 19820115 22903032 24508170 21719571 30249741 21347285 23254933 20798317 21282665
17994087 25244985 20920257 23761445 25348373 12364791 14747013 17210077 24438588 28648823 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24945155 26483478 17510324 20566863 17550304 20353571 25401762 26823975 28004739 19859648 22516182 23537049 15632085 21292972 25474469 26334808 27129103 18362917 19820115 22903032 24508170 21719571 30249741 21347285 23254933 20798317 21282665
EMBL
BABH01031739
NWSH01000005
PCG80993.1
AGBW02014393
OWR41761.1
KQ458863
+ More
KPJ04730.1 JRES01000819 KNC28136.1 GFDF01004063 JAV10021.1 GANO01001768 JAB58103.1 GBXI01010374 JAD03918.1 GGFM01006892 MBW27643.1 GAMC01009490 JAB97065.1 CH933806 EDW13859.1 ADMH02002035 ETN59839.1 GAKP01014276 JAC44676.1 GGFK01002899 MBW36220.1 GGFJ01004065 MBW53206.1 GAMD01002616 JAA98974.1 KA645918 KM215141 AFP60547.1 AIU39614.1 AAAB01008859 EAA44880.4 GEBQ01015285 JAT24692.1 APCN01000533 AXCN02000034 ATLV01017659 ATLV01017660 ATLV01017661 KE525181 KFB42366.1 GDHF01000476 JAI51838.1 GFDG01002341 JAV16458.1 CH940650 EDW66678.1 GECZ01026164 JAS43605.1 CH902617 EDV42833.1 NNAY01001420 OXU24035.1 GFDL01011199 JAV23846.1 AE014297 AY122201 AAF54292.2 AAM52713.1 AXCM01000485 JXUM01138996 JXUM01138997 JXUM01138998 GAPW01000856 KQ568825 JAC12742.1 KXJ68873.1 CH964232 EDW80995.1 AJWK01013934 AJWK01013935 AJWK01013936 AJWK01013937 UFQS01000011 UFQT01000011 SSW97178.1 SSX17564.1 CH477194 EAT48473.1 DS234988 EEB09903.1 GEDC01025784 JAS11514.1 CM000160 EDW96578.2 CH954181 EDV49862.1 CCAG010001926 CCAG010001927 CCAG010001928 EZ422409 ADD18685.1 CP012526 ALC48003.1 GBHO01015250 GBRD01014563 GDHC01011610 JAG28354.1 JAG51263.1 JAQ07019.1 DS231918 EDS26238.1 GEZM01092928 JAV56425.1 CVRI01000064 CRL05343.1 OUUW01000008 SPP84024.1 FJ821033 ACN94734.1 BT127647 KB632375 AEE62609.1 ERL93864.1 CM000070 EAL28975.2 GDIQ01006672 JAN88065.1 LRGB01001361 KZS12267.1 CH479185 EDW38243.1 GL732526 EFX87987.1 GBBI01002287 JAC16425.1 GECL01003477 JAP02647.1 GDIP01032593 JAM71122.1 GBGD01001089 JAC87800.1 GDIP01184479 JAJ38923.1 GFTR01007038 JAW09388.1 GALX01002776 JAB65690.1 KQ979854 KYN18814.1 GDKW01001223 JAI55372.1 KQ971309 EEZ99111.1 ACPB03005390 MH492355 AYA22349.1 HQ668094 AET34921.1 KF621129 AHB33465.1 KQ981522 KYN40705.1 KK107503 EZA49598.1 GDRN01052970 JAI66213.1 KQ978143 KYM96783.1 GL888423 EGI61148.1 KQ982926 KYQ49234.1 QOIP01000006 RLU21896.1 ADTU01026217 NEVH01027073 PNF13845.1 GECZ01030291 JAS39478.1 AMQM01007697 KB097656 ESN92287.1 AF427597 AAL27405.1 GDIP01188779 JAJ34623.1 GL445059 EFN60224.1 GL764659 EFZ17306.1 LBMM01000647 KMQ97894.1 GFWV01002299 MAA27029.1
KPJ04730.1 JRES01000819 KNC28136.1 GFDF01004063 JAV10021.1 GANO01001768 JAB58103.1 GBXI01010374 JAD03918.1 GGFM01006892 MBW27643.1 GAMC01009490 JAB97065.1 CH933806 EDW13859.1 ADMH02002035 ETN59839.1 GAKP01014276 JAC44676.1 GGFK01002899 MBW36220.1 GGFJ01004065 MBW53206.1 GAMD01002616 JAA98974.1 KA645918 KM215141 AFP60547.1 AIU39614.1 AAAB01008859 EAA44880.4 GEBQ01015285 JAT24692.1 APCN01000533 AXCN02000034 ATLV01017659 ATLV01017660 ATLV01017661 KE525181 KFB42366.1 GDHF01000476 JAI51838.1 GFDG01002341 JAV16458.1 CH940650 EDW66678.1 GECZ01026164 JAS43605.1 CH902617 EDV42833.1 NNAY01001420 OXU24035.1 GFDL01011199 JAV23846.1 AE014297 AY122201 AAF54292.2 AAM52713.1 AXCM01000485 JXUM01138996 JXUM01138997 JXUM01138998 GAPW01000856 KQ568825 JAC12742.1 KXJ68873.1 CH964232 EDW80995.1 AJWK01013934 AJWK01013935 AJWK01013936 AJWK01013937 UFQS01000011 UFQT01000011 SSW97178.1 SSX17564.1 CH477194 EAT48473.1 DS234988 EEB09903.1 GEDC01025784 JAS11514.1 CM000160 EDW96578.2 CH954181 EDV49862.1 CCAG010001926 CCAG010001927 CCAG010001928 EZ422409 ADD18685.1 CP012526 ALC48003.1 GBHO01015250 GBRD01014563 GDHC01011610 JAG28354.1 JAG51263.1 JAQ07019.1 DS231918 EDS26238.1 GEZM01092928 JAV56425.1 CVRI01000064 CRL05343.1 OUUW01000008 SPP84024.1 FJ821033 ACN94734.1 BT127647 KB632375 AEE62609.1 ERL93864.1 CM000070 EAL28975.2 GDIQ01006672 JAN88065.1 LRGB01001361 KZS12267.1 CH479185 EDW38243.1 GL732526 EFX87987.1 GBBI01002287 JAC16425.1 GECL01003477 JAP02647.1 GDIP01032593 JAM71122.1 GBGD01001089 JAC87800.1 GDIP01184479 JAJ38923.1 GFTR01007038 JAW09388.1 GALX01002776 JAB65690.1 KQ979854 KYN18814.1 GDKW01001223 JAI55372.1 KQ971309 EEZ99111.1 ACPB03005390 MH492355 AYA22349.1 HQ668094 AET34921.1 KF621129 AHB33465.1 KQ981522 KYN40705.1 KK107503 EZA49598.1 GDRN01052970 JAI66213.1 KQ978143 KYM96783.1 GL888423 EGI61148.1 KQ982926 KYQ49234.1 QOIP01000006 RLU21896.1 ADTU01026217 NEVH01027073 PNF13845.1 GECZ01030291 JAS39478.1 AMQM01007697 KB097656 ESN92287.1 AF427597 AAL27405.1 GDIP01188779 JAJ34623.1 GL445059 EFN60224.1 GL764659 EFZ17306.1 LBMM01000647 KMQ97894.1 GFWV01002299 MAA27029.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053268
UP000037069
UP000075884
+ More
UP000069272 UP000009192 UP000076408 UP000000673 UP000075903 UP000075902 UP000076407 UP000007062 UP000075920 UP000075840 UP000075886 UP000030765 UP000075885 UP000008792 UP000075881 UP000095300 UP000007801 UP000215335 UP000000803 UP000075883 UP000069940 UP000249989 UP000192221 UP000007798 UP000092461 UP000008820 UP000009046 UP000002282 UP000008711 UP000092444 UP000075880 UP000092553 UP000192223 UP000002320 UP000075900 UP000183832 UP000268350 UP000030742 UP000001819 UP000076858 UP000008744 UP000000305 UP000078492 UP000085678 UP000007266 UP000015103 UP000078541 UP000053097 UP000078542 UP000007755 UP000075809 UP000279307 UP000005205 UP000235965 UP000015101 UP000000311 UP000036403
UP000069272 UP000009192 UP000076408 UP000000673 UP000075903 UP000075902 UP000076407 UP000007062 UP000075920 UP000075840 UP000075886 UP000030765 UP000075885 UP000008792 UP000075881 UP000095300 UP000007801 UP000215335 UP000000803 UP000075883 UP000069940 UP000249989 UP000192221 UP000007798 UP000092461 UP000008820 UP000009046 UP000002282 UP000008711 UP000092444 UP000075880 UP000092553 UP000192223 UP000002320 UP000075900 UP000183832 UP000268350 UP000030742 UP000001819 UP000076858 UP000008744 UP000000305 UP000078492 UP000085678 UP000007266 UP000015103 UP000078541 UP000053097 UP000078542 UP000007755 UP000075809 UP000279307 UP000005205 UP000235965 UP000015101 UP000000311 UP000036403
PRIDE
Interpro
IPR027409
GroEL-like_apical_dom_sf
+ More
IPR027413 GROEL-like_equatorial_sf
IPR002194 Chaperonin_TCP-1_CS
IPR012720 Chap_CCT_eta
IPR017998 Chaperone_TCP-1
IPR027410 TCP-1-like_intermed_sf
IPR002423 Cpn60/TCP-1
IPR004210 BESS_motif
IPR006578 MADF-dom
IPR019594 Glu/Gly-bd
IPR036056 Fibrinogen-like_C
IPR014715 Fibrinogen_a/b/g_C_2
IPR014716 Fibrinogen_a/b/g_C_1
IPR002181 Fibrinogen_a/b/g_C_dom
IPR020837 Fibrinogen_CS
IPR027413 GROEL-like_equatorial_sf
IPR002194 Chaperonin_TCP-1_CS
IPR012720 Chap_CCT_eta
IPR017998 Chaperone_TCP-1
IPR027410 TCP-1-like_intermed_sf
IPR002423 Cpn60/TCP-1
IPR004210 BESS_motif
IPR006578 MADF-dom
IPR019594 Glu/Gly-bd
IPR036056 Fibrinogen-like_C
IPR014715 Fibrinogen_a/b/g_C_2
IPR014716 Fibrinogen_a/b/g_C_1
IPR002181 Fibrinogen_a/b/g_C_dom
IPR020837 Fibrinogen_CS
ProteinModelPortal
H9JRM2
A0A2A4KB20
A0A212EJW0
A0A0N1I5L4
A0A0L0C7B5
A0A1L8DUA7
+ More
U5EYW9 A0A1Y9H2B3 A0A0A1WZV3 A0A182F992 A0A2M3ZGM4 W8B7A3 B4KDM5 A0A182Y3V9 W5JAM7 A0A034VMZ5 A0A2M4A661 A0A2M4BJF5 T1DGV8 A0A182UQ34 T1PDA9 A0A182U284 A0A182XIU0 Q7PGM0 A0A182VZP9 A0A1B6LLS6 A0A182HTH9 A0A182QWQ4 A0A084VWM2 A0A0K8WKZ7 A0A182P504 A0A1L8ECI8 B4LWV7 A0A1B6F0B1 A0A182JW05 A0A1I8NV51 B3LXX7 A0A232F081 A0A1Q3F8H6 Q9VHL2 A0A182M116 A0A023EUT4 A0A1W4W2H7 B4NB31 A0A1B0CJ02 A0A336LHQ7 Q17P27 E0V947 A0A1B6CDI9 B4PTW7 B3P4R8 D3TLV7 A0A182J8C4 A0A0M3QYN8 A0A0A9Y855 A0A1W4X638 B0WFH0 A0A182RSY3 A0A1Y1K9S1 A0A1J1IYW5 A0A3B0JP67 D0QWN5 J3JWM9 Q294I9 A0A0P6IC77 A0A164VFC7 B4GL19 E9FX08 A0A023F4G7 A0A0V0G3V3 A0A0P6AN96 A0A069DV50 A0A0P5BTH5 A0A224XHV4 V5G6I1 A0A151J6G8 A0A0P4VTR7 A0A1S3I1C0 D6WBZ5 T1IDR0 A0A385L3J3 J9PNK3 V5RDK5 A0A195FJ69 A0A026W0P5 A0A0P4WFK2 A0A195C9J2 F4WX99 A0A151WMY4 A0A3L8DNN7 A0A158NU49 A0A2J7PBY2 A0A1B6ENH8 T1FNQ7 Q95V46 A0A0P5B6L9 E2B249 E9IQ54 A0A0J7L5X5 A0A293LGP1
U5EYW9 A0A1Y9H2B3 A0A0A1WZV3 A0A182F992 A0A2M3ZGM4 W8B7A3 B4KDM5 A0A182Y3V9 W5JAM7 A0A034VMZ5 A0A2M4A661 A0A2M4BJF5 T1DGV8 A0A182UQ34 T1PDA9 A0A182U284 A0A182XIU0 Q7PGM0 A0A182VZP9 A0A1B6LLS6 A0A182HTH9 A0A182QWQ4 A0A084VWM2 A0A0K8WKZ7 A0A182P504 A0A1L8ECI8 B4LWV7 A0A1B6F0B1 A0A182JW05 A0A1I8NV51 B3LXX7 A0A232F081 A0A1Q3F8H6 Q9VHL2 A0A182M116 A0A023EUT4 A0A1W4W2H7 B4NB31 A0A1B0CJ02 A0A336LHQ7 Q17P27 E0V947 A0A1B6CDI9 B4PTW7 B3P4R8 D3TLV7 A0A182J8C4 A0A0M3QYN8 A0A0A9Y855 A0A1W4X638 B0WFH0 A0A182RSY3 A0A1Y1K9S1 A0A1J1IYW5 A0A3B0JP67 D0QWN5 J3JWM9 Q294I9 A0A0P6IC77 A0A164VFC7 B4GL19 E9FX08 A0A023F4G7 A0A0V0G3V3 A0A0P6AN96 A0A069DV50 A0A0P5BTH5 A0A224XHV4 V5G6I1 A0A151J6G8 A0A0P4VTR7 A0A1S3I1C0 D6WBZ5 T1IDR0 A0A385L3J3 J9PNK3 V5RDK5 A0A195FJ69 A0A026W0P5 A0A0P4WFK2 A0A195C9J2 F4WX99 A0A151WMY4 A0A3L8DNN7 A0A158NU49 A0A2J7PBY2 A0A1B6ENH8 T1FNQ7 Q95V46 A0A0P5B6L9 E2B249 E9IQ54 A0A0J7L5X5 A0A293LGP1
PDB
4B2T
E-value=0,
Score=1814
Ontologies
GO
PANTHER
Topology
Subcellular location
Cytoplasm
Nucleus
Nucleus
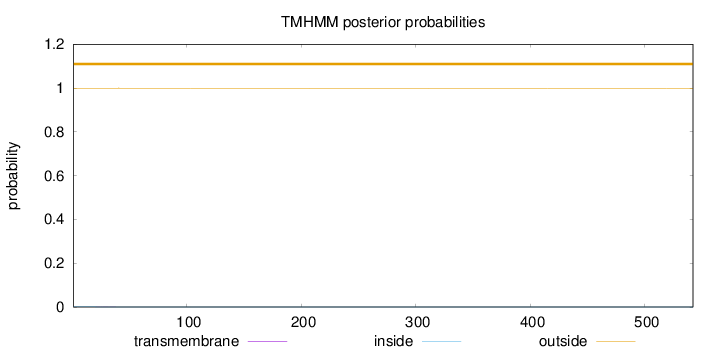
Length:
542
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0038
Exp number, first 60 AAs:
0.00266
Total prob of N-in:
0.00070
outside
1 - 542
Population Genetic Test Statistics
Pi
227.310728
Theta
159.67381
Tajima's D
1.934459
CLR
0.157258
CSRT
0.869306534673266
Interpretation
Uncertain
