Gene
KWMTBOMO14884 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA012171
Annotation
PREDICTED:_spectrin_beta_chain?_non-erythrocytic_5_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.52 Nuclear Reliability : 1.993
Sequence
CDS
ATGCGCGTGGACGACCGCCGCGTGCGCAGCGTGCGGGCGCTCGCCGACAAGCTGCTCAACCAGGGCCCCACCCAGCAGGCGGCCACGGTGGCGCAGCGCCGCGACGCCTTCCTGTCGAAGTGGGCGGCCCTCAGCGGAGCCCTACAGCGGTACCGGGACAACCTGGCCGCCGCCCTCGAGATACATTCGTTTAACAGAGACGTGGAAGATACGGCGGAGCGTATCTCGAAGAAGGCGGCCATCTTCAGCAGCACGGAGCGCGGGCGCGAGCTGGAGGCCGCCCACCTCCTGCGCCGCAGCCACCTGGCGCGCGCCGCCGAGGCCTCCGCCGTGCTCACCAGGATCCAGGAGCTTCAGGCGGACGGCACCGACCTCGCCACCAGACACCCAGAGCACGCTAAAGATATAGAGGAGAGTCTCCGCAACTTACGCGAGGGTTGGGACAACCTGCAGAAGTTGGCTGAGAGGAGAACCGCGCTGCTAGATGCGGCTATCAACGAGCACAAATTCGATGAGAACTTAAAGGAATTGGAAGTCTGGGTGTCGGAAGCCGTGAAGCAGATGGACGAAGTCGAGGACCCGGAGACGATGGCAGAAGCCCAGGCCTTGCTGGAGCTTCACGACGAGAAGAAGGCCGAAATCGACAACCGCCAGAAAGCCATAGCCTCGCTGAAGAAGGAGGCCGAGCAAACGCCGGAGAAGCTTAAGAAAGTCGAACACCTCTCCACCACCCTCGACCAAGCCTGGCTGAAGAGGAAGGAGAATTTGACTCAGGCGCACCAGTTACAGCTACTGAAGGAGCAAGCGAGACAGATTGAAGACTGGCTCTCGACTAAGGAGGCCTTCTTGAACAATGACGACTTGGGAGAGAACTTGGACGCCGTCGAGACCCTCATCCGTAAGCACGCTGAGTTCAGCAAGCTGCTGGAGAGCCAGATCTGCCGCGTCGACGAGCTGAGGAAGAGCGCTGAGGGCTTGAAGGACCATCCTCTGAGGGTCAGGGTGGACGACGTCCTCCGCAGGGCCGACAGACTCAAGGAGTCGTGTAAGTCGCGCGGAGACGTGCTGCAGCAGTCCAGACAGCTGCACCAGTTCCTACAGGACCTGCAGCACGAGAGGGAGTGGATCGCGCTCAAGATGCAGATCGCCAACGACCAGAATTACAGAGAACTGTCTAACCTGCAATCCAAGATACAGAGGCACGCCGCCTTTGAATCAGAGCTTGCGGCCAACAAGGGACGCATCGACGACGTCGCCAACAGAGGGGAGGCGCTTATTGAATCAAAACATTACGCTTCTCAAGAGATAGCGAAACACGTCGAGGAGTTGGAGAACCAGTGGCAGGAATTGCAGACCGCCGCTAAGCTGCGTCACGATCGCCTGCAAGAAGCCTACCAGGCCAGGATCTTCTTCCGAGGGTTAGACGACTTCACCACCTGGCTGGACGAGGTGGAGGCGCAGCTGCTCAGCGAGGACCACGGCAAGGACCTGGCGTCGGTGACGGTCCTTCTGAAGCGACACTCGCGTCTCGAGCAGCAGGTGTCGAGCAAGGCCGACACGGCGACGCAATTGGCCGACACCGCCCGCCAGCTCGCCGACACCAAGCACTTCATGGCACAGGAGATACTGGACAGAGCCGACCAGGCCGTCAAACGCTACCGCCAGCTACAAGAGCCGATACAGATCAGAAGAGATAATCTAGAAGATGCCGAGCTTCTCCACCGCTGGGATAGAGACGCCGACGAGGAATTCGCCTGGTTACAGGAACGGGAGCCCTTGGTGCACCAGGAGGACGCCGGCTCCTCGCTCCCGGAGGCGCAAGCTCTGCTGAAGAAGCACCTGTCTCTGGAAGCTGAGCTTATCGCCAGAGAGCCCACGGTCAGCGCCGTGTGCGGGCGCGCGGCGCAGCTATCCCGGCGAGGTCACTTCGCGTCGGGCGCCCTAGAGGCCCGGGCGCGCGAGTTGTCGGCGCTCATGCGGTCCCTGCTGCACGGCGCCAGCGTGAGGACCGCGGCACTGCAACGACGGTGCGACGTGCTTATGCTGGTGTCGGAGGTAGCGGAGGCGCGCGCGTGGCTGGCGGCGCGGCGGGGGGCGGTGGGGGGCGGGGCGGGGGCCGAGCCGCGGGACGAGGGGGAGGCTCGCGGCCTCACGCGCCGCCTGCACGCCGCGCTCGCAGACCTCACGCCTTTCGACCTCACGCTGGACAAGCTGTACAAGGCCTGCGCCGCTCTGCAGGAGCGCGGCGCGGGGGACGCGGACGAGGTCAAGCGCAGCATCGACGAGCTGAAGGCGGAGCATGAGGACATGAAGCTGCTGGCGGCCAGGAGGCAGCAGCGCCTCGAGCAGAGCGTCAAGTACTTCAAGTTCGTGGAGGAGTGCGAGGAGGTCTCCGAATGGATCGGGGAGCAGATGGCGGCGGCCGCCAGCGAGGACTACGGGCTGGACGTGGAGCACGTAGACACGCTGCAGCAGGCCTTCGCGAGCTTTTTGACTCAACTTAACGCGAACGAGGGTCGCATCGAGTCCGTCTGCGAGGCCGGCAACGTGCTGCTGGAAGAGAACAACCCTGAGTCGGACAAGGTGAAGCAACGGGTCGACGACATCAGGGGTCTGTGGGAGGACCTCAAGGAACTAGCGGTGGCGCGGCAGGAGGCGCTGGCCGGGGCGCGGCAGGTGCACGAGTTCGACCGCAGCGCGGACGAGACGGGCGCGTGGATCGAGGAGAAGGAGCGCGCGCTCAGCGAGCTCAGCCCCAGCGCGCACGCGCAGCCCGCGCACGGACGCGCGCTGCACGCCCTGCGCGCCGACCTGCACGCCGTGCGCGACCAGCACCAACGTCTGCGGGACGAGGCCGCCAGGCTGGGGGAAACGTTCCCTGACGCCCGCGAGCACCTGTCTTCGAAGCTGGAGGACGTGACGGAAGCCCTGGACGCCCTGGAGCGGCGCGCCGAGCACTGCGCGCAGCAGCAGGAGCTGGCCAGCCACCTGCAGGCCTACTTCGGCACCTACCAGGAGCTCCTGGCGTGGTGCAGCGCCACGCTGGCCCGGGTGACGGCGCCCGAGCTGTCCCGGGACGTGCCCGGGGCGGAGCGCCTGGTCGCCAGGCATGACGACATCAAGTCGGAGATCGACTCCAAGGACGAAGACTTCAACAAGTTCTACAGCGACGGCGAAAAGCTGGTCAGAGAAGGCCACTTCATGTCAGCCGAGATAGAGGAGAGGATGTCCGCCCTCCGCAGCCGCCGCGCCGTGCTGGACTCGTGCTGGAGCTCGCGAAGCAGGATCTACGCGCAACACCTGGACGCTCTGGTCTTCAAGCGCGATGCCATGACCCTGGACAACTGGATCACGAATAGAGTGCCGCTGGTGAGGGACGGGAAACTTGGTGAGAACGTGGCCCAAGTTGAGGAGTTGATCACGCGCCACCGAGACCTGGAGGAGACCATCGAGGCGCAGAGGGACAAGTTCAACGCCCTCAAGCGCATCACCCTGATCGAGCAAGCCTTCAAGGAGCAACAAGAAGCGGAGGCGGTGGCGCGTCGCCGGTCCGCCGAGAAGCAAGAGGCAGACCGGCTGCAGCAGTACAGGAGGAGGGAGATGGAACGCATCACCGAGGAGAGGAGGAGGGAAACTATCAGTAGAGAGACGGAGCACGCGCCGACGCAGCGGCCGCCCTCCCCGGAGGAGGGCCTGTCCCCGGCGCCGCAGTTCGACAAGCTGCCCAAGAGCGACGCCGGCGTCAAGCGCGCAGAGAGCATGAACGTCGTGAAGACGCCGAAGAGGACCCCCTCGTTCACGACCCGGCGTCGGACCCAGAGCTTCACCAGGCACCGGGGAGGGGACATCGACAAGCTACCGCCCGTGGAGATCGAAGGCTATTTGGAGCGCAAGCAGCAGGCCGGGTGCGGCGGGAAGCGCGCCACGGTCCGGTCGTGGCGCGGCTACCACGCCGTGCTCTGCGGGCAGCTGCTCTGCTTCTTCAGGGACCAGATGGACTTCGCAAGCACCAAGGCCGCGGCGCCGCCCGTCGCCATACTCAATGCGGAGTGCTCCGCGGCCCCGGACTACACCAAGCGCAGCCACGTGTTCCGGCTCCGCTGCGCCGACGGGGCCGAGTACCTGTTCTCGTGCGGCAGCCGCGCGCTGCTGCAGGACTGGGTCAACAAGATCTCCTTCCACGCGCGGCTGCCCCCGGAGCTGCAGCTGACGCCCTACGCCGCGCCCGGGGACTCGCCCACCGCCGAGATCCGAAGGCGGCTCCAACAAAACGCCTCGTCGTCGTCCGAAGAGGAACTGTCTCCGGAGCCCCAGCGCAAGTCCCGGTCCCAGGCGGAGATCCTGCAGGAGCACAGGAGCAGCCAGCAGGCGGCCAGGAGTAGCGCCACACCAGAGCGGACCACCACCACGACCGAAAGACTCATTGAGTCGACAGTGCTGCCGTCGCTGCCGCCCCGACAGCCGCCGCGCGAGGAGGAGGCCGCCGACGTCGTCCTCAGGAACGCGGAGCAGGCCGCCGGCGGGACCTGGGGCAGGTCCAGATTCTCAAACGGACGAGACATCAATTCAGACTTCATCAGGTCACAGAAAGACGCATACGGAGGTGGTGGAAGCGGTGCCAGGCCGGCGTCCGTGGCTGGTAGCGGAGGCTCTCCCGCCGTAGAGCGGCCCGCCTCGCGGTCGTCGGGCGAGTCGGAGCTGTCCGTGAGCAGTGTGACGAAAGACAAGAAAGAGAAGAAGGGAGTCTTCGGAGGCTTCTTCAGCAAGAAGAAGCGGCCGCAGAGTCACATGTAG
Protein
MRVDDRRVRSVRALADKLLNQGPTQQAATVAQRRDAFLSKWAALSGALQRYRDNLAAALEIHSFNRDVEDTAERISKKAAIFSSTERGRELEAAHLLRRSHLARAAEASAVLTRIQELQADGTDLATRHPEHAKDIEESLRNLREGWDNLQKLAERRTALLDAAINEHKFDENLKELEVWVSEAVKQMDEVEDPETMAEAQALLELHDEKKAEIDNRQKAIASLKKEAEQTPEKLKKVEHLSTTLDQAWLKRKENLTQAHQLQLLKEQARQIEDWLSTKEAFLNNDDLGENLDAVETLIRKHAEFSKLLESQICRVDELRKSAEGLKDHPLRVRVDDVLRRADRLKESCKSRGDVLQQSRQLHQFLQDLQHEREWIALKMQIANDQNYRELSNLQSKIQRHAAFESELAANKGRIDDVANRGEALIESKHYASQEIAKHVEELENQWQELQTAAKLRHDRLQEAYQARIFFRGLDDFTTWLDEVEAQLLSEDHGKDLASVTVLLKRHSRLEQQVSSKADTATQLADTARQLADTKHFMAQEILDRADQAVKRYRQLQEPIQIRRDNLEDAELLHRWDRDADEEFAWLQEREPLVHQEDAGSSLPEAQALLKKHLSLEAELIAREPTVSAVCGRAAQLSRRGHFASGALEARARELSALMRSLLHGASVRTAALQRRCDVLMLVSEVAEARAWLAARRGAVGGGAGAEPRDEGEARGLTRRLHAALADLTPFDLTLDKLYKACAALQERGAGDADEVKRSIDELKAEHEDMKLLAARRQQRLEQSVKYFKFVEECEEVSEWIGEQMAAAASEDYGLDVEHVDTLQQAFASFLTQLNANEGRIESVCEAGNVLLEENNPESDKVKQRVDDIRGLWEDLKELAVARQEALAGARQVHEFDRSADETGAWIEEKERALSELSPSAHAQPAHGRALHALRADLHAVRDQHQRLRDEAARLGETFPDAREHLSSKLEDVTEALDALERRAEHCAQQQELASHLQAYFGTYQELLAWCSATLARVTAPELSRDVPGAERLVARHDDIKSEIDSKDEDFNKFYSDGEKLVREGHFMSAEIEERMSALRSRRAVLDSCWSSRSRIYAQHLDALVFKRDAMTLDNWITNRVPLVRDGKLGENVAQVEELITRHRDLEETIEAQRDKFNALKRITLIEQAFKEQQEAEAVARRRSAEKQEADRLQQYRRREMERITEERRRETISRETEHAPTQRPPSPEEGLSPAPQFDKLPKSDAGVKRAESMNVVKTPKRTPSFTTRRRTQSFTRHRGGDIDKLPPVEIEGYLERKQQAGCGGKRATVRSWRGYHAVLCGQLLCFFRDQMDFASTKAAAPPVAILNAECSAAPDYTKRSHVFRLRCADGAEYLFSCGSRALLQDWVNKISFHARLPPELQLTPYAAPGDSPTAEIRRRLQQNASSSSEEELSPEPQRKSRSQAEILQEHRSSQQAARSSATPERTTTTTERLIESTVLPSLPPRQPPREEEAADVVLRNAEQAAGGTWGRSRFSNGRDINSDFIRSQKDAYGGGGSGARPASVAGSGGSPAVERPASRSSGESELSVSSVTKDKKEKKGVFGGFFSKKKRPQSHM
Summary
Uniprot
H9JRL1
A0A2A4K9M3
A0A2A4KAI8
A0A2H1VZP7
A0A0N1PEQ6
A0A0N0U3D9
+ More
V9IDX7 V9IEH0 A0A087ZZB0 A0A2A3EB80 K7ISG3 A0A232FEG8 A0A0C9RM62 A0A0C9RNA7 A0A0C9RQ97 A0A310SIG7 A0A146LX34 A0A0K8SSY7 A0A2S2NMW7 A0A2H8TWI2 A0A182GHS2 Q176W5 W4VRA2 E2C4R5 W8AJY4 W8B5I6 A0A026WPC2 A0A3L8DWJ0 W8AB01 A0A034V2K7 A0A0K8UTU1 A0A195CSF9 A0A158P0K6 A0A195BN56 A0A151XJJ0 B4HTR1 A0A0J9UDP6 A0A0J9RNN1 A0A0J9RN82 A0A0J9RPN7 A0A0J9RN49 B4PGP9 A0A0R1DV52 A0A0J9RN87 A0A0R1E1I1 A0A0R1DYP9 A0A0J9RPP3 A0A195E0P1 A0A0R1DVK7 A0A0J9RNM4 A0A0R1E094 A0A2R5L953 A0A0J7KVM5 Q7KV70 M9PBL6 A8JNJ6 Q9VZQ3 Q7KV69 A0A0Q5U3S2 A0A0Q5UHY5 A0A0Q5U5L8 B3NBV8 A0A0Q5U3D5 A0A0Q5U9W7 B3M838 A0A0P8XUB3 A0A154P9T1 A0A0P8XUK0 A0A0P8YHA4 A0A0P8XUB9 A0A0L0C3C9 A0A1W4W068 A0A1W4W0Z7 A0A1W4VNW5 A0A1W4W102 A0A1W4VNM6 A0A0L7R381 T1PEG3 T1PEL0 A0A0M4E8S7 A0A2J7PPU1 A0A2J7PPQ2 A0A1I8MT47 A0A2J7PPR5 A0A2J7PPR1 A0A3B0JYR6 A0A2J7PPR7 A0A2J7PPQ1 A0A2J7PPS9 A0A1I8MT55 A0A2J7PPS2 A0A1I8MT52 A0A3B0KMZ6 A0A3B0K3F9 A0A3B0K1L9 A0A3B0KPU3 D6WA70 A0A2J7PPS8 A0A1I8MT51
V9IDX7 V9IEH0 A0A087ZZB0 A0A2A3EB80 K7ISG3 A0A232FEG8 A0A0C9RM62 A0A0C9RNA7 A0A0C9RQ97 A0A310SIG7 A0A146LX34 A0A0K8SSY7 A0A2S2NMW7 A0A2H8TWI2 A0A182GHS2 Q176W5 W4VRA2 E2C4R5 W8AJY4 W8B5I6 A0A026WPC2 A0A3L8DWJ0 W8AB01 A0A034V2K7 A0A0K8UTU1 A0A195CSF9 A0A158P0K6 A0A195BN56 A0A151XJJ0 B4HTR1 A0A0J9UDP6 A0A0J9RNN1 A0A0J9RN82 A0A0J9RPN7 A0A0J9RN49 B4PGP9 A0A0R1DV52 A0A0J9RN87 A0A0R1E1I1 A0A0R1DYP9 A0A0J9RPP3 A0A195E0P1 A0A0R1DVK7 A0A0J9RNM4 A0A0R1E094 A0A2R5L953 A0A0J7KVM5 Q7KV70 M9PBL6 A8JNJ6 Q9VZQ3 Q7KV69 A0A0Q5U3S2 A0A0Q5UHY5 A0A0Q5U5L8 B3NBV8 A0A0Q5U3D5 A0A0Q5U9W7 B3M838 A0A0P8XUB3 A0A154P9T1 A0A0P8XUK0 A0A0P8YHA4 A0A0P8XUB9 A0A0L0C3C9 A0A1W4W068 A0A1W4W0Z7 A0A1W4VNW5 A0A1W4W102 A0A1W4VNM6 A0A0L7R381 T1PEG3 T1PEL0 A0A0M4E8S7 A0A2J7PPU1 A0A2J7PPQ2 A0A1I8MT47 A0A2J7PPR5 A0A2J7PPR1 A0A3B0JYR6 A0A2J7PPR7 A0A2J7PPQ1 A0A2J7PPS9 A0A1I8MT55 A0A2J7PPS2 A0A1I8MT52 A0A3B0KMZ6 A0A3B0K3F9 A0A3B0K1L9 A0A3B0KPU3 D6WA70 A0A2J7PPS8 A0A1I8MT51
Pubmed
EMBL
BABH01031739
BABH01031740
BABH01031741
BABH01031742
BABH01031743
BABH01031744
+ More
BABH01031745 BABH01031746 BABH01031747 BABH01031748 NWSH01000005 PCG80995.1 PCG80996.1 ODYU01005424 SOQ46253.1 KQ458863 KPJ04733.1 KQ435885 KOX69621.1 JR039834 AEY58867.1 JR039833 AEY58866.1 KZ288295 PBC28978.1 NNAY01000364 OXU28868.1 GBYB01009365 JAG79132.1 GBYB01009830 JAG79597.1 GBYB01009366 JAG79133.1 KQ768090 OAD53242.1 GDHC01006421 JAQ12208.1 GBRD01009523 JAG56301.1 GGMR01005673 MBY18292.1 GFXV01006801 MBW18606.1 JXUM01064422 JXUM01064423 JXUM01064424 JXUM01064425 KQ562297 KXJ76213.1 CH477381 EAT42207.1 GANO01003796 JAB56075.1 GL452568 EFN77073.1 GAMC01021547 JAB85008.1 GAMC01021546 JAB85009.1 KK107139 EZA57818.1 QOIP01000003 RLU24811.1 GAMC01021545 JAB85010.1 GAKP01022575 JAC36377.1 GDHF01022195 GDHF01003466 JAI30119.1 JAI48848.1 KQ977394 KYN03044.1 ADTU01005248 ADTU01005249 ADTU01005250 ADTU01005251 KQ976440 KYM86614.1 KQ982074 KYQ60465.1 CH480817 EDW50332.1 CM002912 KMY97350.1 KMY97352.1 KMY97348.1 KMY97355.1 KMY97349.1 KMY97346.1 CM000159 EDW93267.2 KRK01062.1 KMY97351.1 KMY97353.1 KRK01058.1 KRK01063.1 KRK01059.1 KRK01060.1 KMY97354.1 KQ979955 KYN18507.1 KRK01061.1 KMY97347.1 KRK01057.1 GGLE01001873 MBY05999.1 LBMM01002799 KMQ94348.1 AE014296 AAS64957.1 AGB94063.1 AGB94062.1 ABW08452.2 AAF47766.1 ADV37478.1 AAS64958.1 CH954178 KQS43499.1 KQS43498.1 KQS43500.1 EDV50775.2 KQS43502.1 KQS43503.1 KQS43501.1 CH902618 EDV39946.2 KPU78310.1 KQ434839 KZC07978.1 KPU78313.1 KPU78314.1 KPU78311.1 KPU78312.1 JRES01001060 KNC25934.1 KQ414663 KOC65284.1 KA647177 AFP61806.1 KA647176 AFP61805.1 CP012525 ALC43341.1 NEVH01022647 PNF18326.1 PNF18328.1 PNF18333.1 PNF18330.1 OUUW01000012 SPP87215.1 PNF18334.1 PNF18327.1 PNF18331.1 PNF18332.1 SPP87216.1 SPP87212.1 SPP87213.1 SPP87211.1 KQ971312 EEZ98593.1 PNF18335.1
BABH01031745 BABH01031746 BABH01031747 BABH01031748 NWSH01000005 PCG80995.1 PCG80996.1 ODYU01005424 SOQ46253.1 KQ458863 KPJ04733.1 KQ435885 KOX69621.1 JR039834 AEY58867.1 JR039833 AEY58866.1 KZ288295 PBC28978.1 NNAY01000364 OXU28868.1 GBYB01009365 JAG79132.1 GBYB01009830 JAG79597.1 GBYB01009366 JAG79133.1 KQ768090 OAD53242.1 GDHC01006421 JAQ12208.1 GBRD01009523 JAG56301.1 GGMR01005673 MBY18292.1 GFXV01006801 MBW18606.1 JXUM01064422 JXUM01064423 JXUM01064424 JXUM01064425 KQ562297 KXJ76213.1 CH477381 EAT42207.1 GANO01003796 JAB56075.1 GL452568 EFN77073.1 GAMC01021547 JAB85008.1 GAMC01021546 JAB85009.1 KK107139 EZA57818.1 QOIP01000003 RLU24811.1 GAMC01021545 JAB85010.1 GAKP01022575 JAC36377.1 GDHF01022195 GDHF01003466 JAI30119.1 JAI48848.1 KQ977394 KYN03044.1 ADTU01005248 ADTU01005249 ADTU01005250 ADTU01005251 KQ976440 KYM86614.1 KQ982074 KYQ60465.1 CH480817 EDW50332.1 CM002912 KMY97350.1 KMY97352.1 KMY97348.1 KMY97355.1 KMY97349.1 KMY97346.1 CM000159 EDW93267.2 KRK01062.1 KMY97351.1 KMY97353.1 KRK01058.1 KRK01063.1 KRK01059.1 KRK01060.1 KMY97354.1 KQ979955 KYN18507.1 KRK01061.1 KMY97347.1 KRK01057.1 GGLE01001873 MBY05999.1 LBMM01002799 KMQ94348.1 AE014296 AAS64957.1 AGB94063.1 AGB94062.1 ABW08452.2 AAF47766.1 ADV37478.1 AAS64958.1 CH954178 KQS43499.1 KQS43498.1 KQS43500.1 EDV50775.2 KQS43502.1 KQS43503.1 KQS43501.1 CH902618 EDV39946.2 KPU78310.1 KQ434839 KZC07978.1 KPU78313.1 KPU78314.1 KPU78311.1 KPU78312.1 JRES01001060 KNC25934.1 KQ414663 KOC65284.1 KA647177 AFP61806.1 KA647176 AFP61805.1 CP012525 ALC43341.1 NEVH01022647 PNF18326.1 PNF18328.1 PNF18333.1 PNF18330.1 OUUW01000012 SPP87215.1 PNF18334.1 PNF18327.1 PNF18331.1 PNF18332.1 SPP87216.1 SPP87212.1 SPP87213.1 SPP87211.1 KQ971312 EEZ98593.1 PNF18335.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000053105
UP000005203
UP000242457
+ More
UP000002358 UP000215335 UP000069940 UP000249989 UP000008820 UP000008237 UP000053097 UP000279307 UP000078542 UP000005205 UP000078540 UP000075809 UP000001292 UP000002282 UP000078492 UP000036403 UP000000803 UP000008711 UP000007801 UP000076502 UP000037069 UP000192221 UP000053825 UP000092553 UP000235965 UP000095301 UP000268350 UP000007266
UP000002358 UP000215335 UP000069940 UP000249989 UP000008820 UP000008237 UP000053097 UP000279307 UP000078542 UP000005205 UP000078540 UP000075809 UP000001292 UP000002282 UP000078492 UP000036403 UP000000803 UP000008711 UP000007801 UP000076502 UP000037069 UP000192221 UP000053825 UP000092553 UP000235965 UP000095301 UP000268350 UP000007266
Interpro
Gene 3D
CDD
ProteinModelPortal
H9JRL1
A0A2A4K9M3
A0A2A4KAI8
A0A2H1VZP7
A0A0N1PEQ6
A0A0N0U3D9
+ More
V9IDX7 V9IEH0 A0A087ZZB0 A0A2A3EB80 K7ISG3 A0A232FEG8 A0A0C9RM62 A0A0C9RNA7 A0A0C9RQ97 A0A310SIG7 A0A146LX34 A0A0K8SSY7 A0A2S2NMW7 A0A2H8TWI2 A0A182GHS2 Q176W5 W4VRA2 E2C4R5 W8AJY4 W8B5I6 A0A026WPC2 A0A3L8DWJ0 W8AB01 A0A034V2K7 A0A0K8UTU1 A0A195CSF9 A0A158P0K6 A0A195BN56 A0A151XJJ0 B4HTR1 A0A0J9UDP6 A0A0J9RNN1 A0A0J9RN82 A0A0J9RPN7 A0A0J9RN49 B4PGP9 A0A0R1DV52 A0A0J9RN87 A0A0R1E1I1 A0A0R1DYP9 A0A0J9RPP3 A0A195E0P1 A0A0R1DVK7 A0A0J9RNM4 A0A0R1E094 A0A2R5L953 A0A0J7KVM5 Q7KV70 M9PBL6 A8JNJ6 Q9VZQ3 Q7KV69 A0A0Q5U3S2 A0A0Q5UHY5 A0A0Q5U5L8 B3NBV8 A0A0Q5U3D5 A0A0Q5U9W7 B3M838 A0A0P8XUB3 A0A154P9T1 A0A0P8XUK0 A0A0P8YHA4 A0A0P8XUB9 A0A0L0C3C9 A0A1W4W068 A0A1W4W0Z7 A0A1W4VNW5 A0A1W4W102 A0A1W4VNM6 A0A0L7R381 T1PEG3 T1PEL0 A0A0M4E8S7 A0A2J7PPU1 A0A2J7PPQ2 A0A1I8MT47 A0A2J7PPR5 A0A2J7PPR1 A0A3B0JYR6 A0A2J7PPR7 A0A2J7PPQ1 A0A2J7PPS9 A0A1I8MT55 A0A2J7PPS2 A0A1I8MT52 A0A3B0KMZ6 A0A3B0K3F9 A0A3B0K1L9 A0A3B0KPU3 D6WA70 A0A2J7PPS8 A0A1I8MT51
V9IDX7 V9IEH0 A0A087ZZB0 A0A2A3EB80 K7ISG3 A0A232FEG8 A0A0C9RM62 A0A0C9RNA7 A0A0C9RQ97 A0A310SIG7 A0A146LX34 A0A0K8SSY7 A0A2S2NMW7 A0A2H8TWI2 A0A182GHS2 Q176W5 W4VRA2 E2C4R5 W8AJY4 W8B5I6 A0A026WPC2 A0A3L8DWJ0 W8AB01 A0A034V2K7 A0A0K8UTU1 A0A195CSF9 A0A158P0K6 A0A195BN56 A0A151XJJ0 B4HTR1 A0A0J9UDP6 A0A0J9RNN1 A0A0J9RN82 A0A0J9RPN7 A0A0J9RN49 B4PGP9 A0A0R1DV52 A0A0J9RN87 A0A0R1E1I1 A0A0R1DYP9 A0A0J9RPP3 A0A195E0P1 A0A0R1DVK7 A0A0J9RNM4 A0A0R1E094 A0A2R5L953 A0A0J7KVM5 Q7KV70 M9PBL6 A8JNJ6 Q9VZQ3 Q7KV69 A0A0Q5U3S2 A0A0Q5UHY5 A0A0Q5U5L8 B3NBV8 A0A0Q5U3D5 A0A0Q5U9W7 B3M838 A0A0P8XUB3 A0A154P9T1 A0A0P8XUK0 A0A0P8YHA4 A0A0P8XUB9 A0A0L0C3C9 A0A1W4W068 A0A1W4W0Z7 A0A1W4VNW5 A0A1W4W102 A0A1W4VNM6 A0A0L7R381 T1PEG3 T1PEL0 A0A0M4E8S7 A0A2J7PPU1 A0A2J7PPQ2 A0A1I8MT47 A0A2J7PPR5 A0A2J7PPR1 A0A3B0JYR6 A0A2J7PPR7 A0A2J7PPQ1 A0A2J7PPS9 A0A1I8MT55 A0A2J7PPS2 A0A1I8MT52 A0A3B0KMZ6 A0A3B0K3F9 A0A3B0K1L9 A0A3B0KPU3 D6WA70 A0A2J7PPS8 A0A1I8MT51
PDB
1U4Q
E-value=3.91787e-27,
Score=308
Ontologies
GO
GO:0003779
GO:0005543
GO:0010797
GO:0042060
GO:0005912
GO:0032432
GO:0008017
GO:0006897
GO:0016324
GO:0032509
GO:0016327
GO:0045186
GO:0005886
GO:0008092
GO:0030707
GO:0007009
GO:0008091
GO:0030036
GO:0005815
GO:0031941
GO:0005515
GO:0006508
GO:0003676
GO:0016701
GO:0006355
GO:0003824
GO:0008152
GO:0016491
GO:0015930
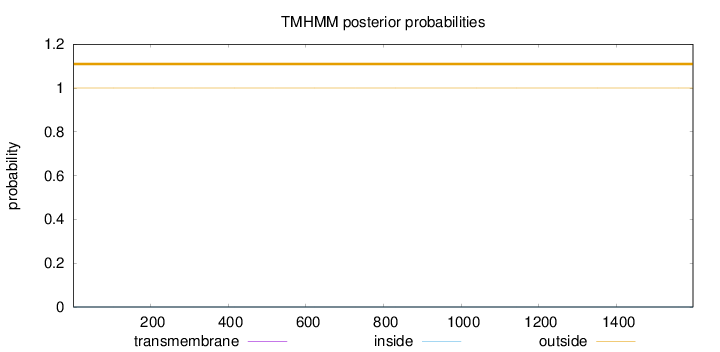
Topology
Length:
1597
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00153
Exp number, first 60 AAs:
0.001
Total prob of N-in:
0.00007
outside
1 - 1597
Population Genetic Test Statistics
Pi
242.158201
Theta
160.053613
Tajima's D
1.286487
CLR
0.40034
CSRT
0.729563521823909
Interpretation
Uncertain
