Gene
KWMTBOMO14874
Pre Gene Modal
BGIBMGA012186
Annotation
PREDICTED:_leucine-rich_repeat_serine/threonine-protein_kinase_1_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.12 Mitochondrial Reliability : 1.175
Sequence
CDS
ATGAAGATCGAGGACTTAGCGCATGTCTTCAAAGCTTCACCCATGTTAGCGCGCGGGGAGGAGGCCAGCTCCTTAGGCGTCTCGCTACTGAACAAGTTCGAGCTGGCCCTCTGCTGGGATTCCAGGGTCTTACTGGTCCCACCGTTGCTGCCGAATGCGGAGCCGGCGAGCCCTCAGCTGGCTTTACGCAGCAAGACCTGGTCACATTCGAGCAGGAACACTCCGAGGCGAGCCTTCCCTGCGTCCGTGCAGCCCCACCTCACCCTTCCGGAACAGAGCAGGCCGACCGATTCCCCAGTGTTGATACAAGATCACACGGGCTCCATCACGGTGTCTGGGAGAGGGGGGGAGATGTCACTTGTCCGACTCGTCCTGCTCTCGTACGTGCCGAGTGGCTTCTGGGCTCGTCTGACCACCAGGGTGCTGGCCGATGCTGCACTGGCAGTGGCTGCGGGCCAACTGTATCGTCTAACACCCGAGATGGAGGTGGATGAGGGTATATCCCGAGCGCTGGAGGGATGCTGGGGCTGGAAGCTGTGGAAAACGGGACTGTCGCTGTGTTGCGGGGCTCTCACCGTCGCCTCGCTGCGGGACCTGCCGGCGCGAACCGACGCCCTGCTCGGCCTCTGCGACGACGGACACACTGAACACGCTTACACGGGACTGAGGCTCAGGGTACGTCAAGAAGGTGCTTGGTGTGATCTGGAGGTGGACGCGACTGCCTGCGTGGAGCTCTGTCTACCCGCGCAGGTGTGTGTCGTGAAGAGAGACGACGGCCTCCCGATAGCTGGGTATCAGAGCATCAGCCTGGAACCGAACCCAGAGACGTTGGCCAAACTGCTAGCGCTACTTTCTGACCACGTGGACTTGTTACTAGAAGACTGGTACCCATCCTTAGGTACAAGGTTCGTACATACTTCGGAGGGACGATGCCTGATCACGCGGCTGGTCCCCTGCCCCGCCTGTCTGAAGGACGCGGGGCGAAGCTCCCACGACGACATGCCTTTAGCGCAAGCATTCAGAAGATTTGAACTTGGCGCCTTGGAGCCGAGGAGGGCGCGGCTGTCCCAGGACTCCCGCGCCTCAGACGGAGACAGTGGAGTTGGCGCTGAGTCTAATGCCTCGTCCCGGCTCGGGTCAGTGGAGGGGATCGTGAACGAGCCAAGACCCGCCACCTCCGTGTACGCTTGGACCGTGGAGGACTGCATATTAGCGGCTTGCACCAGTACTAAGCTCCAGTGCGCGTTGCATTCGCACGTAGAGCTGAGGCAGTTGGCTCCGGATGCGCTCCTGCTAGACGTGGAGGAAGAGAAGAGAGCCCGGTGGGAGCAAATCACGCGCGGGGTGGTGGCGGGGCGCGGCGCCTTCGGTACCGTGCTGACGGGCACGTGGCGCCGTTCGCCAGCTAACTTAGTGCCGGTCGCCATCAAAGCCTTACAGCCAGTCCCGCCCCCCAGTCACCATGATGTACCAGCTCAGCAGGCTTACAAGTACCCCATCTCGCGACGCGCTATCGACGCGTCACTTTCGGAGGGTCCCTTGATCTGTATGTTACCGTAG
Protein
MKIEDLAHVFKASPMLARGEEASSLGVSLLNKFELALCWDSRVLLVPPLLPNAEPASPQLALRSKTWSHSSRNTPRRAFPASVQPHLTLPEQSRPTDSPVLIQDHTGSITVSGRGGEMSLVRLVLLSYVPSGFWARLTTRVLADAALAVAAGQLYRLTPEMEVDEGISRALEGCWGWKLWKTGLSLCCGALTVASLRDLPARTDALLGLCDDGHTEHAYTGLRLRVRQEGAWCDLEVDATACVELCLPAQVCVVKRDDGLPIAGYQSISLEPNPETLAKLLALLSDHVDLLLEDWYPSLGTRFVHTSEGRCLITRLVPCPACLKDAGRSSHDDMPLAQAFRRFELGALEPRRARLSQDSRASDGDSGVGAESNASSRLGSVEGIVNEPRPATSVYAWTVEDCILAACTSTKLQCALHSHVELRQLAPDALLLDVEEEKRARWEQITRGVVAGRGAFGTVLTGTWRRSPANLVPVAIKALQPVPPPSHHDVPAQQAYKYPISRRAIDASLSEGPLICMLP
Summary
Uniprot
H9JRM6
A0A2A4KB68
A0A2A4KAR1
A0A0N0PEE2
A0A0N0PAG7
A0A3S2LWW4
+ More
A0A232F2H4 A0A1W4WRZ4 A0A1W4WSZ2 A0A1B6DQ53 A0A3Q0IZP8 E0VNV3 A0A023F2Y8 A0A067RI47 A0A0P6JRT1 A0A0C9R2L5 A0A182GWL1 A0A088A8J7 A0A2A3E2R1 A0A224X4P3 A0A1B0CX03 A0A2R7WA29 A0A336M2J0 A0A026WKY3 F4WG42 A0A3L8D628 A0A1Y1MLJ9 A0A158NT23 A0A0J7KXP6 A0A195BHG2 A0A195F0J5 D6WA18 E2BN57 E2AKZ5 E9IBF0 A0A0P5UWM1 A0A0P5UZY3 A0A0P6CXY4 A0A0P5F8L3 A0A1J1HTA3 A0A0P5WAQ0 A0A0P4XVS5 A0A0P5SQB3 A0A0N8CKL0 A0A0N8BCM2 A0A0P5HX33 A0A0N8AZU8 A0A0P6G6X9 A0A0N7ZMR0 A0A0P5WWD9 A0A0P5SPY2 A0A0P5B7Q8 A0A0P5RIG2 A0A0P5IRB8 A0A0P4ZM56 A0A0P6HGI5 A0A0P6FL56 A0A0N8AVG1 A0A0P5KND3 A0A0N8C006 A0A0P5ZYU7 A0A0P6BX16 A0A0P5QKI8 A0A0P5KJL2 A0A0N8E9S2 A0A0P5M3Y1 A0A0P6EXZ9 A0A0N8BKQ8 A0A0N7ZM35 A0A0P5DYY4 A0A0P5YU81 A0A0P6E2G8 A0A0P6IB48 A0A0P5D7P1 A0A0P4YIT6 A0A0P8ZZT0 B3M0A7 T1HIN4 T1IVJ9 A0A164KNT3 A0A2L2Y445 A0A0A9XQE6 B0WZ06 A0A195C8P3 A0A087SVZ3 A0A151XBT5 A0A3B0KMK8 A0A3B0KHA4 A0A154NZP0 X1WSM3 A0A2S2Q8Y9 B4II95 A0A1A9WY73 B4PPQ9 A0A0R1E0Y7 A0A0P5SU68 A0A1A9Z8E1 A0A1W4V9W5
A0A232F2H4 A0A1W4WRZ4 A0A1W4WSZ2 A0A1B6DQ53 A0A3Q0IZP8 E0VNV3 A0A023F2Y8 A0A067RI47 A0A0P6JRT1 A0A0C9R2L5 A0A182GWL1 A0A088A8J7 A0A2A3E2R1 A0A224X4P3 A0A1B0CX03 A0A2R7WA29 A0A336M2J0 A0A026WKY3 F4WG42 A0A3L8D628 A0A1Y1MLJ9 A0A158NT23 A0A0J7KXP6 A0A195BHG2 A0A195F0J5 D6WA18 E2BN57 E2AKZ5 E9IBF0 A0A0P5UWM1 A0A0P5UZY3 A0A0P6CXY4 A0A0P5F8L3 A0A1J1HTA3 A0A0P5WAQ0 A0A0P4XVS5 A0A0P5SQB3 A0A0N8CKL0 A0A0N8BCM2 A0A0P5HX33 A0A0N8AZU8 A0A0P6G6X9 A0A0N7ZMR0 A0A0P5WWD9 A0A0P5SPY2 A0A0P5B7Q8 A0A0P5RIG2 A0A0P5IRB8 A0A0P4ZM56 A0A0P6HGI5 A0A0P6FL56 A0A0N8AVG1 A0A0P5KND3 A0A0N8C006 A0A0P5ZYU7 A0A0P6BX16 A0A0P5QKI8 A0A0P5KJL2 A0A0N8E9S2 A0A0P5M3Y1 A0A0P6EXZ9 A0A0N8BKQ8 A0A0N7ZM35 A0A0P5DYY4 A0A0P5YU81 A0A0P6E2G8 A0A0P6IB48 A0A0P5D7P1 A0A0P4YIT6 A0A0P8ZZT0 B3M0A7 T1HIN4 T1IVJ9 A0A164KNT3 A0A2L2Y445 A0A0A9XQE6 B0WZ06 A0A195C8P3 A0A087SVZ3 A0A151XBT5 A0A3B0KMK8 A0A3B0KHA4 A0A154NZP0 X1WSM3 A0A2S2Q8Y9 B4II95 A0A1A9WY73 B4PPQ9 A0A0R1E0Y7 A0A0P5SU68 A0A1A9Z8E1 A0A1W4V9W5
Pubmed
EMBL
BABH01031763
BABH01031764
BABH01031765
BABH01031766
BABH01031767
BABH01031768
+ More
BABH01031769 BABH01031770 BABH01031771 NWSH01000005 PCG80990.1 PCG80988.1 KQ459988 KPJ18810.1 KQ458863 KPJ04736.1 RSAL01000140 RVE46091.1 NNAY01001222 OXU24690.1 GEDC01009496 JAS27802.1 DS235354 EEB15059.1 GBBI01002997 JAC15715.1 KK852667 KDR18932.1 GDUN01001016 JAN94903.1 GBYB01001096 JAG70863.1 JXUM01093607 KQ564068 KXJ72845.1 KZ288419 PBC25995.1 GFTR01008960 JAW07466.1 AJWK01033084 KK854504 PTY16409.1 UFQT01000309 SSX23103.1 KK107185 EZA55764.1 GL888128 EGI66809.1 QOIP01000012 RLU15689.1 GEZM01033860 JAV84077.1 ADTU01025480 LBMM01002255 KMQ95071.1 KQ976488 KYM83606.1 KQ981897 KYN33677.1 KQ971312 EEZ98574.2 GL449385 EFN82819.1 GL440425 EFN65912.1 GL762111 EFZ22114.1 GDIP01107617 JAL96097.1 GDIP01107618 JAL96096.1 GDIQ01086023 JAN08714.1 GDIQ01266297 JAJ85427.1 CVRI01000014 CRK89702.1 GDIP01088772 JAM14943.1 GDIP01237306 JAI86095.1 GDIP01136865 JAL66849.1 GDIP01122567 JAL81147.1 GDIQ01190884 GDIQ01190883 JAK60842.1 GDIQ01226672 JAK25053.1 GDIQ01226675 GDIQ01226674 GDIQ01226673 JAK25050.1 GDIQ01049041 JAN45696.1 GDIP01230257 JAI93144.1 GDIP01092672 JAM11043.1 GDIQ01101118 JAL50608.1 GDIP01189158 JAJ34244.1 GDIQ01101120 GDIQ01101119 GDIQ01101117 JAL50606.1 GDIQ01209079 JAK42646.1 GDIP01215007 JAJ08395.1 GDIQ01033096 JAN61641.1 GDIQ01047891 JAN46846.1 GDIQ01238971 JAK12754.1 GDIQ01181536 JAK70189.1 GDIQ01128211 JAL23515.1 GDIP01041333 GDIP01007018 JAM62382.1 GDIP01008316 JAM95399.1 GDIQ01113412 JAL38314.1 GDIQ01184689 JAK67036.1 GDIQ01047892 JAN46845.1 GDIQ01161245 JAK90480.1 GDIQ01068834 GDIQ01068833 GDIQ01068832 GDIQ01068831 JAN25906.1 GDIQ01168195 JAK83530.1 GDIP01232057 JAI91344.1 GDIP01167109 JAJ56293.1 GDIP01053378 JAM50337.1 GDIQ01068779 JAN25958.1 GDIQ01007064 JAN87673.1 GDIP01161762 JAJ61640.1 GDIP01230258 JAI93143.1 CH902617 KPU80090.1 EDV43113.2 ACPB03012567 JH431584 LRGB01003275 KZS03423.1 IAAA01005380 LAA02080.1 GBHO01024279 JAG19325.1 DS232202 EDS37320.1 KQ978205 KYM96488.1 KK112200 KFM57032.1 KQ982320 KYQ57809.1 OUUW01000013 SPP87819.1 SPP87820.1 KQ434782 KZC04554.1 ABLF02032569 GGMS01004888 MBY74091.1 CH480842 EDW49639.1 CM000160 EDW96159.2 KRK02944.1 GDIP01142565 JAL61149.1
BABH01031769 BABH01031770 BABH01031771 NWSH01000005 PCG80990.1 PCG80988.1 KQ459988 KPJ18810.1 KQ458863 KPJ04736.1 RSAL01000140 RVE46091.1 NNAY01001222 OXU24690.1 GEDC01009496 JAS27802.1 DS235354 EEB15059.1 GBBI01002997 JAC15715.1 KK852667 KDR18932.1 GDUN01001016 JAN94903.1 GBYB01001096 JAG70863.1 JXUM01093607 KQ564068 KXJ72845.1 KZ288419 PBC25995.1 GFTR01008960 JAW07466.1 AJWK01033084 KK854504 PTY16409.1 UFQT01000309 SSX23103.1 KK107185 EZA55764.1 GL888128 EGI66809.1 QOIP01000012 RLU15689.1 GEZM01033860 JAV84077.1 ADTU01025480 LBMM01002255 KMQ95071.1 KQ976488 KYM83606.1 KQ981897 KYN33677.1 KQ971312 EEZ98574.2 GL449385 EFN82819.1 GL440425 EFN65912.1 GL762111 EFZ22114.1 GDIP01107617 JAL96097.1 GDIP01107618 JAL96096.1 GDIQ01086023 JAN08714.1 GDIQ01266297 JAJ85427.1 CVRI01000014 CRK89702.1 GDIP01088772 JAM14943.1 GDIP01237306 JAI86095.1 GDIP01136865 JAL66849.1 GDIP01122567 JAL81147.1 GDIQ01190884 GDIQ01190883 JAK60842.1 GDIQ01226672 JAK25053.1 GDIQ01226675 GDIQ01226674 GDIQ01226673 JAK25050.1 GDIQ01049041 JAN45696.1 GDIP01230257 JAI93144.1 GDIP01092672 JAM11043.1 GDIQ01101118 JAL50608.1 GDIP01189158 JAJ34244.1 GDIQ01101120 GDIQ01101119 GDIQ01101117 JAL50606.1 GDIQ01209079 JAK42646.1 GDIP01215007 JAJ08395.1 GDIQ01033096 JAN61641.1 GDIQ01047891 JAN46846.1 GDIQ01238971 JAK12754.1 GDIQ01181536 JAK70189.1 GDIQ01128211 JAL23515.1 GDIP01041333 GDIP01007018 JAM62382.1 GDIP01008316 JAM95399.1 GDIQ01113412 JAL38314.1 GDIQ01184689 JAK67036.1 GDIQ01047892 JAN46845.1 GDIQ01161245 JAK90480.1 GDIQ01068834 GDIQ01068833 GDIQ01068832 GDIQ01068831 JAN25906.1 GDIQ01168195 JAK83530.1 GDIP01232057 JAI91344.1 GDIP01167109 JAJ56293.1 GDIP01053378 JAM50337.1 GDIQ01068779 JAN25958.1 GDIQ01007064 JAN87673.1 GDIP01161762 JAJ61640.1 GDIP01230258 JAI93143.1 CH902617 KPU80090.1 EDV43113.2 ACPB03012567 JH431584 LRGB01003275 KZS03423.1 IAAA01005380 LAA02080.1 GBHO01024279 JAG19325.1 DS232202 EDS37320.1 KQ978205 KYM96488.1 KK112200 KFM57032.1 KQ982320 KYQ57809.1 OUUW01000013 SPP87819.1 SPP87820.1 KQ434782 KZC04554.1 ABLF02032569 GGMS01004888 MBY74091.1 CH480842 EDW49639.1 CM000160 EDW96159.2 KRK02944.1 GDIP01142565 JAL61149.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000283053
UP000215335
+ More
UP000192223 UP000079169 UP000009046 UP000027135 UP000069940 UP000249989 UP000005203 UP000242457 UP000092461 UP000053097 UP000007755 UP000279307 UP000005205 UP000036403 UP000078540 UP000078541 UP000007266 UP000008237 UP000000311 UP000183832 UP000007801 UP000015103 UP000076858 UP000002320 UP000078542 UP000054359 UP000075809 UP000268350 UP000076502 UP000007819 UP000001292 UP000091820 UP000002282 UP000092445 UP000192221
UP000192223 UP000079169 UP000009046 UP000027135 UP000069940 UP000249989 UP000005203 UP000242457 UP000092461 UP000053097 UP000007755 UP000279307 UP000005205 UP000036403 UP000078540 UP000078541 UP000007266 UP000008237 UP000000311 UP000183832 UP000007801 UP000015103 UP000076858 UP000002320 UP000078542 UP000054359 UP000075809 UP000268350 UP000076502 UP000007819 UP000001292 UP000091820 UP000002282 UP000092445 UP000192221
PRIDE
Pfam
Interpro
IPR020683
Ankyrin_rpt-contain_dom
+ More
IPR020859 ROC_dom
IPR036770 Ankyrin_rpt-contain_sf
IPR032171 COR
IPR003591 Leu-rich_rpt_typical-subtyp
IPR000719 Prot_kinase_dom
IPR011047 Quinoprotein_ADH-like_supfam
IPR001611 Leu-rich_rpt
IPR027417 P-loop_NTPase
IPR032675 LRR_dom_sf
IPR011009 Kinase-like_dom_sf
IPR002110 Ankyrin_rpt
IPR008271 Ser/Thr_kinase_AS
IPR006434 Pyrimidine_nucleotidase_eu
IPR023214 HAD_sf
IPR036412 HAD-like_sf
IPR036322 WD40_repeat_dom_sf
IPR017441 Protein_kinase_ATP_BS
IPR025875 Leu-rich_rpt_4
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR007497 DUF541
IPR020859 ROC_dom
IPR036770 Ankyrin_rpt-contain_sf
IPR032171 COR
IPR003591 Leu-rich_rpt_typical-subtyp
IPR000719 Prot_kinase_dom
IPR011047 Quinoprotein_ADH-like_supfam
IPR001611 Leu-rich_rpt
IPR027417 P-loop_NTPase
IPR032675 LRR_dom_sf
IPR011009 Kinase-like_dom_sf
IPR002110 Ankyrin_rpt
IPR008271 Ser/Thr_kinase_AS
IPR006434 Pyrimidine_nucleotidase_eu
IPR023214 HAD_sf
IPR036412 HAD-like_sf
IPR036322 WD40_repeat_dom_sf
IPR017441 Protein_kinase_ATP_BS
IPR025875 Leu-rich_rpt_4
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR007497 DUF541
SUPFAM
Gene 3D
CDD
ProteinModelPortal
H9JRM6
A0A2A4KB68
A0A2A4KAR1
A0A0N0PEE2
A0A0N0PAG7
A0A3S2LWW4
+ More
A0A232F2H4 A0A1W4WRZ4 A0A1W4WSZ2 A0A1B6DQ53 A0A3Q0IZP8 E0VNV3 A0A023F2Y8 A0A067RI47 A0A0P6JRT1 A0A0C9R2L5 A0A182GWL1 A0A088A8J7 A0A2A3E2R1 A0A224X4P3 A0A1B0CX03 A0A2R7WA29 A0A336M2J0 A0A026WKY3 F4WG42 A0A3L8D628 A0A1Y1MLJ9 A0A158NT23 A0A0J7KXP6 A0A195BHG2 A0A195F0J5 D6WA18 E2BN57 E2AKZ5 E9IBF0 A0A0P5UWM1 A0A0P5UZY3 A0A0P6CXY4 A0A0P5F8L3 A0A1J1HTA3 A0A0P5WAQ0 A0A0P4XVS5 A0A0P5SQB3 A0A0N8CKL0 A0A0N8BCM2 A0A0P5HX33 A0A0N8AZU8 A0A0P6G6X9 A0A0N7ZMR0 A0A0P5WWD9 A0A0P5SPY2 A0A0P5B7Q8 A0A0P5RIG2 A0A0P5IRB8 A0A0P4ZM56 A0A0P6HGI5 A0A0P6FL56 A0A0N8AVG1 A0A0P5KND3 A0A0N8C006 A0A0P5ZYU7 A0A0P6BX16 A0A0P5QKI8 A0A0P5KJL2 A0A0N8E9S2 A0A0P5M3Y1 A0A0P6EXZ9 A0A0N8BKQ8 A0A0N7ZM35 A0A0P5DYY4 A0A0P5YU81 A0A0P6E2G8 A0A0P6IB48 A0A0P5D7P1 A0A0P4YIT6 A0A0P8ZZT0 B3M0A7 T1HIN4 T1IVJ9 A0A164KNT3 A0A2L2Y445 A0A0A9XQE6 B0WZ06 A0A195C8P3 A0A087SVZ3 A0A151XBT5 A0A3B0KMK8 A0A3B0KHA4 A0A154NZP0 X1WSM3 A0A2S2Q8Y9 B4II95 A0A1A9WY73 B4PPQ9 A0A0R1E0Y7 A0A0P5SU68 A0A1A9Z8E1 A0A1W4V9W5
A0A232F2H4 A0A1W4WRZ4 A0A1W4WSZ2 A0A1B6DQ53 A0A3Q0IZP8 E0VNV3 A0A023F2Y8 A0A067RI47 A0A0P6JRT1 A0A0C9R2L5 A0A182GWL1 A0A088A8J7 A0A2A3E2R1 A0A224X4P3 A0A1B0CX03 A0A2R7WA29 A0A336M2J0 A0A026WKY3 F4WG42 A0A3L8D628 A0A1Y1MLJ9 A0A158NT23 A0A0J7KXP6 A0A195BHG2 A0A195F0J5 D6WA18 E2BN57 E2AKZ5 E9IBF0 A0A0P5UWM1 A0A0P5UZY3 A0A0P6CXY4 A0A0P5F8L3 A0A1J1HTA3 A0A0P5WAQ0 A0A0P4XVS5 A0A0P5SQB3 A0A0N8CKL0 A0A0N8BCM2 A0A0P5HX33 A0A0N8AZU8 A0A0P6G6X9 A0A0N7ZMR0 A0A0P5WWD9 A0A0P5SPY2 A0A0P5B7Q8 A0A0P5RIG2 A0A0P5IRB8 A0A0P4ZM56 A0A0P6HGI5 A0A0P6FL56 A0A0N8AVG1 A0A0P5KND3 A0A0N8C006 A0A0P5ZYU7 A0A0P6BX16 A0A0P5QKI8 A0A0P5KJL2 A0A0N8E9S2 A0A0P5M3Y1 A0A0P6EXZ9 A0A0N8BKQ8 A0A0N7ZM35 A0A0P5DYY4 A0A0P5YU81 A0A0P6E2G8 A0A0P6IB48 A0A0P5D7P1 A0A0P4YIT6 A0A0P8ZZT0 B3M0A7 T1HIN4 T1IVJ9 A0A164KNT3 A0A2L2Y445 A0A0A9XQE6 B0WZ06 A0A195C8P3 A0A087SVZ3 A0A151XBT5 A0A3B0KMK8 A0A3B0KHA4 A0A154NZP0 X1WSM3 A0A2S2Q8Y9 B4II95 A0A1A9WY73 B4PPQ9 A0A0R1E0Y7 A0A0P5SU68 A0A1A9Z8E1 A0A1W4V9W5
Ontologies
GO
GO:0004672
GO:0005524
GO:0000287
GO:0008253
GO:0005737
GO:0016301
GO:0016021
GO:1990033
GO:0071212
GO:0051683
GO:0044297
GO:0005795
GO:0005765
GO:0008088
GO:0031902
GO:0004674
GO:0017137
GO:0043198
GO:0040011
GO:1900242
GO:0006914
GO:0050808
GO:0050774
GO:0018105
GO:0048488
GO:0032418
GO:0042147
GO:0005829
GO:1901215
Topology
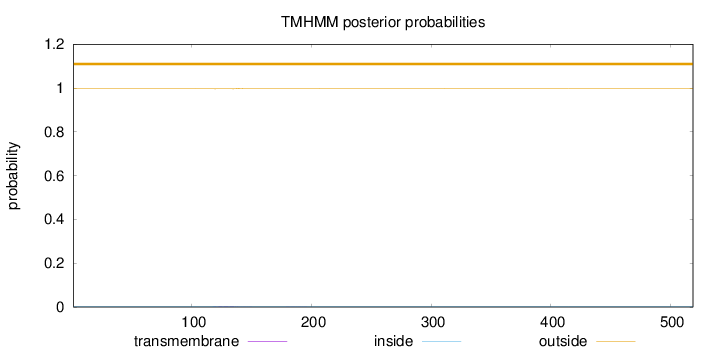
Length:
519
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.05436
Exp number, first 60 AAs:
0.00023
Total prob of N-in:
0.00352
outside
1 - 519
Population Genetic Test Statistics
Pi
244.282063
Theta
161.221345
Tajima's D
1.406518
CLR
0.04186
CSRT
0.766711664416779
Interpretation
Uncertain
