Gene
KWMTBOMO14870 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA012166
Annotation
putative_WD_repeat_domain_34_[Operophtera_brumata]
Location in the cell
Extracellular Reliability : 1.544
Sequence
CDS
ATGTGGAATTTGAGTGGATATGATAGCGAAGCGGTCGGATTCGATTCTGTGGCTGTTAAAAAAACCGAAGACGCGAGCACCGGCACTCAAACTACAGAATTCTTTGAAGCGGCCGTCGCAACTCAAACTGTTGCTTACAAAGACGGTTCATCGATGACCTCTCCTACAGAGGATTTGACGACAAAAGACGTAAAATATTCGGCAGCCAATTTAAATGCGTTCCTCCAAAGAGTAATGCCGGGTATGCTTGAGCAGCTAGACGAGAACAAAGAAAACGCATACGAGTCTTCGGAATCTGACGAGGATAATGAAATTTTGACAGCTAAATTGGTACAAGAGCTAAAGGTACGAGAAGTGCAAGGCAGCGGTGATCAGGCACTGTCTGTACTAGACCTATCTTGGAGTAATACCGGCAACTCGTTAGCCGTGTCTCTAGGACAGAAGCAGCACGAGAATTGGTGCACAGATAACGGTTTAATAAGGTTCTATACGGTCAAAGTCAGTGCCGGCGACAAATTAGTGCACAGCATGGATATCAACGAGAAAAACTGTGTTTCAGTATTGAAATATCATCCGTCGGTCGCGGCTCTGTTCGTCTACGGGACGGTTACGGGCGAGATTGTGATCTGCGACCTTAGAAATAACGATGAACTTCAAATGACGTCGCCGTCTGGTACGCACGGTTCGAAAAGGGTCACGGCGCTTTCTTGGCTGGAGTCCACAACCGCTGAAACGTTTTTTGCGATCCACATCAACAATACAGGCAAGCGAAGGGCCTCGACCGATCGCATACTAGTTTCATCCGGTAGCGACGGAACAATCGTCGCATGGCACGTCAACGCTGACAACAAAACGTTTGAAGCCATTGTAACCTACTCCGTGAACGGAGCCCGCAAGATGGCCGCCCCGGATATCAGCTGTTTTGATTTCATCAACACGTTTCCGCTACGTCCGACGGGCGATAAAATTTCGAACGACATTTTCGTGGTCGGCACGAATGTCGGTAATTTATTTTTGTGTAATATAAAGAACGCTCAGAGCGCAGCGCATAGCGGCCACGTTGATCCGGTCAGCGATGTTCTAGAAAGCCACGCGACCTGCATTTTGGCTGTTACGTTTCACCAAGATAAGCCTGGCGTTTTTATAACACTGTCCATTGATTCCGAATTGAGGATATATAATGTTAAACAGGCCAGTCCCTTGAAGGTGATTTTGCAGTAA
Protein
MWNLSGYDSEAVGFDSVAVKKTEDASTGTQTTEFFEAAVATQTVAYKDGSSMTSPTEDLTTKDVKYSAANLNAFLQRVMPGMLEQLDENKENAYESSESDEDNEILTAKLVQELKVREVQGSGDQALSVLDLSWSNTGNSLAVSLGQKQHENWCTDNGLIRFYTVKVSAGDKLVHSMDINEKNCVSVLKYHPSVAALFVYGTVTGEIVICDLRNNDELQMTSPSGTHGSKRVTALSWLESTTAETFFAIHINNTGKRRASTDRILVSSGSDGTIVAWHVNADNKTFEAIVTYSVNGARKMAAPDISCFDFINTFPLRPTGDKISNDIFVVGTNVGNLFLCNIKNAQSAAHSGHVDPVSDVLESHATCILAVTFHQDKPGVFITLSIDSELRIYNVKQASPLKVILQ
Summary
Uniprot
H9JRK6
A0A0L7LTN1
A0A2H1V0N3
A0A2H1VB68
A0A2W1BH17
A0A2A4J725
+ More
A0A2W1BAI2 A0A212EQ12 A0A0N1PGG3 A0A0N0PE25 A0A067QLH3 A0A2J7PGA3 A0A0L7RJH3 A0A1B6H2D7 K1R5F1 A0A1B6J4Y6 E9IZE3 A0A1W4WBI6 A0A195AU54 A0A1B6KFN5 E2BE55 A0A151WW63 A0A088ADF5 A0A195CXV0 A0A151J3A4 E2AYY3 A0A158P0V5 A0A1B6C6Q1 W4XZX2 A0A210R242 A0A2A3EF88 A0A2C9K504 A0A369RSF3 K7JTB0 A0A151K011 F4WD63 T1IQJ7 A0A0M8ZZE5 A0A026WFJ7 V4A149 C3ZGK1 A0A2J7PGA4 A0A154PNQ4 A0A1S3JBW9 A0A2G8KLK2 A0A0B6ZEV5 A0A075A6H2 B3S2X2 A0A2T7PAM8 A0A061RH73 A0A224XMC4 A0A0B6ZEN6 A0A3S3PDS4 A0A023F792 A7RZ65 A0A1W2WMF6 F7A027 A0A1Y2B9R5 A0A2B4RGB8 A0A1Y2C1L9 A0A0P4VUG9 R4G873 T1KJD3 C1N0M5 R7TIB5 A0A0L8GP68 A0A0K8SAE5 A0A0A9XD44 A0A2P6MX94 A0A091V9A6 A0A091STK9 H3B3P3 A0A1S8VZQ4 A0A146L241 A0A091TXH6 A0A1Y1HJ38 A0A094KBM5 A0A091NK25 A0A2P6KS44 A0A3Q0K2W3 A0A3Q3QVM3 A0A3Q0K5F3 A0A0L0HQL4 A0A091L2A3 A0A3Q3NB38 W5UAC6 A0A2D0SZE4 A0A2R6X6N6 G0UKK9 U3JFB8 D2UY63 T2M823 A0A2D0SYV4 A0A3B3SZI3 A0A093R493 A0A060XS85 A0A3P8Z053
A0A2W1BAI2 A0A212EQ12 A0A0N1PGG3 A0A0N0PE25 A0A067QLH3 A0A2J7PGA3 A0A0L7RJH3 A0A1B6H2D7 K1R5F1 A0A1B6J4Y6 E9IZE3 A0A1W4WBI6 A0A195AU54 A0A1B6KFN5 E2BE55 A0A151WW63 A0A088ADF5 A0A195CXV0 A0A151J3A4 E2AYY3 A0A158P0V5 A0A1B6C6Q1 W4XZX2 A0A210R242 A0A2A3EF88 A0A2C9K504 A0A369RSF3 K7JTB0 A0A151K011 F4WD63 T1IQJ7 A0A0M8ZZE5 A0A026WFJ7 V4A149 C3ZGK1 A0A2J7PGA4 A0A154PNQ4 A0A1S3JBW9 A0A2G8KLK2 A0A0B6ZEV5 A0A075A6H2 B3S2X2 A0A2T7PAM8 A0A061RH73 A0A224XMC4 A0A0B6ZEN6 A0A3S3PDS4 A0A023F792 A7RZ65 A0A1W2WMF6 F7A027 A0A1Y2B9R5 A0A2B4RGB8 A0A1Y2C1L9 A0A0P4VUG9 R4G873 T1KJD3 C1N0M5 R7TIB5 A0A0L8GP68 A0A0K8SAE5 A0A0A9XD44 A0A2P6MX94 A0A091V9A6 A0A091STK9 H3B3P3 A0A1S8VZQ4 A0A146L241 A0A091TXH6 A0A1Y1HJ38 A0A094KBM5 A0A091NK25 A0A2P6KS44 A0A3Q0K2W3 A0A3Q3QVM3 A0A3Q0K5F3 A0A0L0HQL4 A0A091L2A3 A0A3Q3NB38 W5UAC6 A0A2D0SZE4 A0A2R6X6N6 G0UKK9 U3JFB8 D2UY63 T2M823 A0A2D0SYV4 A0A3B3SZI3 A0A093R493 A0A060XS85 A0A3P8Z053
Pubmed
19121390
26227816
28756777
22118469
26354079
24845553
+ More
22992520 21282665 20798317 21347285 28812685 15562597 30042472 20075255 21719571 24508170 30249741 23254933 18563158 29023486 18719581 25474469 17615350 12481130 15114417 27129103 19359590 25401762 29378020 9215903 26823975 24865297 23127152 22331916 20211133 24065732 29240929 24755649 25069045
22992520 21282665 20798317 21347285 28812685 15562597 30042472 20075255 21719571 24508170 30249741 23254933 18563158 29023486 18719581 25474469 17615350 12481130 15114417 27129103 19359590 25401762 29378020 9215903 26823975 24865297 23127152 22331916 20211133 24065732 29240929 24755649 25069045
EMBL
BABH01031771
JTDY01000147
KOB78596.1
ODYU01000137
SOQ34405.1
ODYU01001599
+ More
SOQ38046.1 KZ150412 PZC70973.1 NWSH01002937 PCG67250.1 PZC70974.1 AGBW02013337 OWR43585.1 KQ458863 KPJ04710.1 KQ459988 KPJ18779.1 KK853289 KDR08919.1 NEVH01025635 PNF15363.1 KQ414581 KOC71025.1 GECZ01000952 JAS68817.1 JH816363 EKC40953.1 GECU01013472 JAS94234.1 GL767121 EFZ14057.1 KQ976738 KYM75706.1 GEBQ01029742 JAT10235.1 GL447755 EFN86042.1 KQ982691 KYQ52100.1 KQ977141 KYN05407.1 KQ980300 KYN16811.1 GL444028 EFN61353.1 ADTU01005842 GEDC01028115 JAS09183.1 AAGJ04158193 AAGJ04158194 AAGJ04158195 AAGJ04158196 NEDP02000770 OWF55059.1 KZ288271 PBC29816.1 NOWV01000268 RDD37162.1 AAZX01000105 KQ981414 KYN41890.1 GL888084 EGI67871.1 AFFK01018331 KQ435812 KOX72733.1 KK107235 QOIP01000009 EZA54890.1 RLU18605.1 KB202444 ESO90367.1 GG666619 EEN48337.1 PNF15362.1 KQ434980 KZC13094.1 MRZV01000500 PIK48818.1 HACG01020198 CEK67063.1 KL596621 KER33962.1 DS985248 EDV22864.1 PZQS01000005 PVD30466.1 GBEZ01016124 JAC70099.1 GFTR01006836 JAW09590.1 HACG01020199 CEK67064.1 NCKU01001065 RWS13203.1 GBBI01001609 JAC17103.1 DS469556 EDO43231.1 MCGO01000076 ORY31504.1 LSMT01000662 PFX15302.1 MCGO01000033 ORY40911.1 GDKW01000404 JAI56191.1 GAHY01001472 JAA76038.1 CAEY01000119 GG663744 EEH54298.1 AMQN01002615 KB309694 ELT93583.1 KQ420929 KOF78816.1 GBRD01015707 JAG50119.1 GBHO01045524 GBHO01026881 GBHO01026880 GBHO01026879 JAF98079.1 JAG16723.1 JAG16724.1 JAG16725.1 MDYQ01000337 PRP76276.1 KK733774 KFQ99369.1 KK486152 KFQ62037.1 AFYH01037965 AFYH01037966 AFYH01037967 AFYH01037968 AFYH01037969 LYON01000438 OON07203.1 GDHC01016005 GDHC01009297 JAQ02624.1 JAQ09332.1 KK467950 KFQ82961.1 DF236963 GAQ78524.1 KL347518 KFZ56773.1 KL384538 KFP89432.1 MWRG01006129 PRD29160.1 KQ257452 KND03084.1 KK764264 KFP45878.1 JT410051 AHH38913.1 KZ772705 PTQ41764.1 HE575316 CCC89914.1 AGTO01010820 GG738845 EFC50738.1 HAAD01001885 CDG68117.1 KL224938 KFW65854.1 FR905495 CDQ79735.1
SOQ38046.1 KZ150412 PZC70973.1 NWSH01002937 PCG67250.1 PZC70974.1 AGBW02013337 OWR43585.1 KQ458863 KPJ04710.1 KQ459988 KPJ18779.1 KK853289 KDR08919.1 NEVH01025635 PNF15363.1 KQ414581 KOC71025.1 GECZ01000952 JAS68817.1 JH816363 EKC40953.1 GECU01013472 JAS94234.1 GL767121 EFZ14057.1 KQ976738 KYM75706.1 GEBQ01029742 JAT10235.1 GL447755 EFN86042.1 KQ982691 KYQ52100.1 KQ977141 KYN05407.1 KQ980300 KYN16811.1 GL444028 EFN61353.1 ADTU01005842 GEDC01028115 JAS09183.1 AAGJ04158193 AAGJ04158194 AAGJ04158195 AAGJ04158196 NEDP02000770 OWF55059.1 KZ288271 PBC29816.1 NOWV01000268 RDD37162.1 AAZX01000105 KQ981414 KYN41890.1 GL888084 EGI67871.1 AFFK01018331 KQ435812 KOX72733.1 KK107235 QOIP01000009 EZA54890.1 RLU18605.1 KB202444 ESO90367.1 GG666619 EEN48337.1 PNF15362.1 KQ434980 KZC13094.1 MRZV01000500 PIK48818.1 HACG01020198 CEK67063.1 KL596621 KER33962.1 DS985248 EDV22864.1 PZQS01000005 PVD30466.1 GBEZ01016124 JAC70099.1 GFTR01006836 JAW09590.1 HACG01020199 CEK67064.1 NCKU01001065 RWS13203.1 GBBI01001609 JAC17103.1 DS469556 EDO43231.1 MCGO01000076 ORY31504.1 LSMT01000662 PFX15302.1 MCGO01000033 ORY40911.1 GDKW01000404 JAI56191.1 GAHY01001472 JAA76038.1 CAEY01000119 GG663744 EEH54298.1 AMQN01002615 KB309694 ELT93583.1 KQ420929 KOF78816.1 GBRD01015707 JAG50119.1 GBHO01045524 GBHO01026881 GBHO01026880 GBHO01026879 JAF98079.1 JAG16723.1 JAG16724.1 JAG16725.1 MDYQ01000337 PRP76276.1 KK733774 KFQ99369.1 KK486152 KFQ62037.1 AFYH01037965 AFYH01037966 AFYH01037967 AFYH01037968 AFYH01037969 LYON01000438 OON07203.1 GDHC01016005 GDHC01009297 JAQ02624.1 JAQ09332.1 KK467950 KFQ82961.1 DF236963 GAQ78524.1 KL347518 KFZ56773.1 KL384538 KFP89432.1 MWRG01006129 PRD29160.1 KQ257452 KND03084.1 KK764264 KFP45878.1 JT410051 AHH38913.1 KZ772705 PTQ41764.1 HE575316 CCC89914.1 AGTO01010820 GG738845 EFC50738.1 HAAD01001885 CDG68117.1 KL224938 KFW65854.1 FR905495 CDQ79735.1
Proteomes
UP000005204
UP000037510
UP000218220
UP000007151
UP000053268
UP000053240
+ More
UP000027135 UP000235965 UP000053825 UP000005408 UP000192223 UP000078540 UP000008237 UP000075809 UP000005203 UP000078542 UP000078492 UP000000311 UP000005205 UP000007110 UP000242188 UP000242457 UP000076420 UP000253843 UP000002358 UP000078541 UP000007755 UP000053105 UP000053097 UP000279307 UP000030746 UP000001554 UP000076502 UP000085678 UP000230750 UP000009022 UP000245119 UP000285301 UP000001593 UP000008144 UP000193642 UP000225706 UP000015104 UP000001876 UP000014760 UP000053454 UP000241769 UP000053605 UP000008672 UP000189906 UP000054558 UP000261600 UP000053201 UP000261640 UP000221080 UP000016665 UP000006671 UP000261540 UP000054081 UP000193380 UP000265140
UP000027135 UP000235965 UP000053825 UP000005408 UP000192223 UP000078540 UP000008237 UP000075809 UP000005203 UP000078542 UP000078492 UP000000311 UP000005205 UP000007110 UP000242188 UP000242457 UP000076420 UP000253843 UP000002358 UP000078541 UP000007755 UP000053105 UP000053097 UP000279307 UP000030746 UP000001554 UP000076502 UP000085678 UP000230750 UP000009022 UP000245119 UP000285301 UP000001593 UP000008144 UP000193642 UP000225706 UP000015104 UP000001876 UP000014760 UP000053454 UP000241769 UP000053605 UP000008672 UP000189906 UP000054558 UP000261600 UP000053201 UP000261640 UP000221080 UP000016665 UP000006671 UP000261540 UP000054081 UP000193380 UP000265140
PRIDE
Pfam
PF00400 WD40
Interpro
SUPFAM
SSF50978
SSF50978
Gene 3D
ProteinModelPortal
H9JRK6
A0A0L7LTN1
A0A2H1V0N3
A0A2H1VB68
A0A2W1BH17
A0A2A4J725
+ More
A0A2W1BAI2 A0A212EQ12 A0A0N1PGG3 A0A0N0PE25 A0A067QLH3 A0A2J7PGA3 A0A0L7RJH3 A0A1B6H2D7 K1R5F1 A0A1B6J4Y6 E9IZE3 A0A1W4WBI6 A0A195AU54 A0A1B6KFN5 E2BE55 A0A151WW63 A0A088ADF5 A0A195CXV0 A0A151J3A4 E2AYY3 A0A158P0V5 A0A1B6C6Q1 W4XZX2 A0A210R242 A0A2A3EF88 A0A2C9K504 A0A369RSF3 K7JTB0 A0A151K011 F4WD63 T1IQJ7 A0A0M8ZZE5 A0A026WFJ7 V4A149 C3ZGK1 A0A2J7PGA4 A0A154PNQ4 A0A1S3JBW9 A0A2G8KLK2 A0A0B6ZEV5 A0A075A6H2 B3S2X2 A0A2T7PAM8 A0A061RH73 A0A224XMC4 A0A0B6ZEN6 A0A3S3PDS4 A0A023F792 A7RZ65 A0A1W2WMF6 F7A027 A0A1Y2B9R5 A0A2B4RGB8 A0A1Y2C1L9 A0A0P4VUG9 R4G873 T1KJD3 C1N0M5 R7TIB5 A0A0L8GP68 A0A0K8SAE5 A0A0A9XD44 A0A2P6MX94 A0A091V9A6 A0A091STK9 H3B3P3 A0A1S8VZQ4 A0A146L241 A0A091TXH6 A0A1Y1HJ38 A0A094KBM5 A0A091NK25 A0A2P6KS44 A0A3Q0K2W3 A0A3Q3QVM3 A0A3Q0K5F3 A0A0L0HQL4 A0A091L2A3 A0A3Q3NB38 W5UAC6 A0A2D0SZE4 A0A2R6X6N6 G0UKK9 U3JFB8 D2UY63 T2M823 A0A2D0SYV4 A0A3B3SZI3 A0A093R493 A0A060XS85 A0A3P8Z053
A0A2W1BAI2 A0A212EQ12 A0A0N1PGG3 A0A0N0PE25 A0A067QLH3 A0A2J7PGA3 A0A0L7RJH3 A0A1B6H2D7 K1R5F1 A0A1B6J4Y6 E9IZE3 A0A1W4WBI6 A0A195AU54 A0A1B6KFN5 E2BE55 A0A151WW63 A0A088ADF5 A0A195CXV0 A0A151J3A4 E2AYY3 A0A158P0V5 A0A1B6C6Q1 W4XZX2 A0A210R242 A0A2A3EF88 A0A2C9K504 A0A369RSF3 K7JTB0 A0A151K011 F4WD63 T1IQJ7 A0A0M8ZZE5 A0A026WFJ7 V4A149 C3ZGK1 A0A2J7PGA4 A0A154PNQ4 A0A1S3JBW9 A0A2G8KLK2 A0A0B6ZEV5 A0A075A6H2 B3S2X2 A0A2T7PAM8 A0A061RH73 A0A224XMC4 A0A0B6ZEN6 A0A3S3PDS4 A0A023F792 A7RZ65 A0A1W2WMF6 F7A027 A0A1Y2B9R5 A0A2B4RGB8 A0A1Y2C1L9 A0A0P4VUG9 R4G873 T1KJD3 C1N0M5 R7TIB5 A0A0L8GP68 A0A0K8SAE5 A0A0A9XD44 A0A2P6MX94 A0A091V9A6 A0A091STK9 H3B3P3 A0A1S8VZQ4 A0A146L241 A0A091TXH6 A0A1Y1HJ38 A0A094KBM5 A0A091NK25 A0A2P6KS44 A0A3Q0K2W3 A0A3Q3QVM3 A0A3Q0K5F3 A0A0L0HQL4 A0A091L2A3 A0A3Q3NB38 W5UAC6 A0A2D0SZE4 A0A2R6X6N6 G0UKK9 U3JFB8 D2UY63 T2M823 A0A2D0SYV4 A0A3B3SZI3 A0A093R493 A0A060XS85 A0A3P8Z053
PDB
6F3A
E-value=0.0015354,
Score=99
Ontologies
KEGG
GO
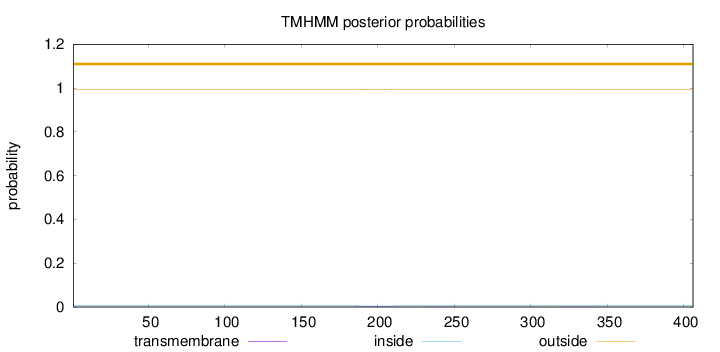
Topology
Length:
406
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0797399999999999
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00672
outside
1 - 406
Population Genetic Test Statistics
Pi
225.469453
Theta
181.685683
Tajima's D
0.704115
CLR
0
CSRT
0.579821008949553
Interpretation
Uncertain
