Pre Gene Modal
BGIBMGA012188
Annotation
PREDICTED:_UDP-glucose_6-dehydrogenase_[Papilio_polytes]
Full name
UDP-glucose 6-dehydrogenase
Alternative Name
Protein sugarless
Protein suppenkasper
Protein suppenkasper
Location in the cell
Cytoplasmic Reliability : 1.779
Sequence
CDS
ATGGTGATACAAAAAATCTGCTGTTTAGGAGCTGGTTATGTCGGCGGTCCAACCTGCAGTGTTATAGCACTGAAATGCCCCAATATTAAGGTAACGGTTTGCGACAAAAGCGAAGAGAGGATAAAGCAATGGAACTCTGATAAGCTACCGATTTACGAGCCTGGCCTAGACGATGTAGTCCGCGAATGTAGAGGTAAAAATTTATTTTTCTCAACGAACATCGCCGAAAGCATTCGGGAAGCCGATTTAATATTTATATCCGTTAACACGCCCACGAAGACAATAGGTAATGGGAAAGGGCGAGCTGCAGACTTGAAGTACATAGAGAGCGCAGCCCGCATGATAGCGGATCTGGCCACCAGCAATAAGATTGTCGTCGAGAAATCGACTGTTCCAGTCAAAGCTGCTGAAATTATTATGAAAATCCTTCGTGCCAATACCAAACCTGGCGTTGAGTACCAGATCCTGTCGAACCCTGAGTTTTTAGCAGAGGGAACCGCTATAGTTGATCTTGTTGAGGCTGAAAGAGTTCTGATCGGCGGGGAAGACACCCCAGAAGGCCAGAAAGCTGTTCAGGAACTATGCTGGGTGTACGAACACTGGATCCCGGCGAAGAACATCCTGACAACCAATACTTGGAGTTCAGAGCTGTCTAAACTTGCAGCAAATGCCTTCCTTGCCCAACGAATATCGAGCATCAACTCGTTATCAGCTGTCTGTGAAGCAACAGGTGCTGATGTGTCAGAAGTGGCCAGAGCTGTTGGTAGGGACTCTCGTATTGGCCCCAAGTTCCTTGAGGCTTCCATCGGCTTTGGGGGCAGTTGTTTCCAGAAAGATATATTAAATTTGATTTATTTGTGTGAATGCTTGAATCTGCCAGAAGTGGCAGCCTACTGGCAGCAAGTGGTGAGCTTAAACGACTACCAGAAGACCAGGTTCACTCGGAAAGTAATTGAGGCACTCTTCAATACAGTATCAGATAAGAAAATAGCTATACTCGGATTCTCATTCAAGAAGAACACAGGGGACACAAGAGAGTCTCCTGCCATATATGTCTGCAGTACGTTATTAGACGAAGGTGCAAAGCTGCACATTTATGATCCGAAGGTGGAGCACGACCAGATATACTATGAGCTGACCAATCCCTATGTTACCTCAGAGCCTGAGAAAGTGAGTTTTTTTCTCTTTTTTGAAGTACTTATTATTTTAGATTATTTTTTTATAAAGAATTTGGCTCCAATGCCTTAG
Protein
MVIQKICCLGAGYVGGPTCSVIALKCPNIKVTVCDKSEERIKQWNSDKLPIYEPGLDDVVRECRGKNLFFSTNIAESIREADLIFISVNTPTKTIGNGKGRAADLKYIESAARMIADLATSNKIVVEKSTVPVKAAEIIMKILRANTKPGVEYQILSNPEFLAEGTAIVDLVEAERVLIGGEDTPEGQKAVQELCWVYEHWIPAKNILTTNTWSSELSKLAANAFLAQRISSINSLSAVCEATGADVSEVARAVGRDSRIGPKFLEASIGFGGSCFQKDILNLIYLCECLNLPEVAAYWQQVVSLNDYQKTRFTRKVIEALFNTVSDKKIAILGFSFKKNTGDTRESPAIYVCSTLLDEGAKLHIYDPKVEHDQIYYELTNPYVTSEPEKVSFFLFFEVLIILDYFFIKNLAPMP
Summary
Description
Involved in the biosynthesis of glycosaminoglycans; hyaluronan, chondroitin sulfate, and heparan sulfate.
Involved in the biosynthesis of glycosaminoglycans; hyaluronan, chondroitin sulfate and heparan sulfate. Required for wingless signaling in different tissues.
Involved in the biosynthesis of glycosaminoglycans; hyaluronan, chondroitin sulfate and heparan sulfate. Required for wingless signaling in different tissues.
Catalytic Activity
H2O + 2 NAD(+) + UDP-alpha-D-glucose = 3 H(+) + 2 NADH + UDP-alpha-D-glucuronate
Similarity
Belongs to the UDP-glucose/GDP-mannose dehydrogenase family.
Keywords
Complete proteome
NAD
Oxidoreductase
Reference proteome
Wnt signaling pathway
Feature
chain UDP-glucose 6-dehydrogenase
Uniprot
H9JRM8
A0A0N1I5K8
A0A0N1IQ45
A0A2A4J5K4
A0A2H1VCB3
A0A2W1B998
+ More
A0A212EYQ8 A0A0L7L164 A0A069DTZ0 A0A023F4G4 T1HCB1 K7J7N2 E2AU70 A0A088A2I2 A0A0P4VSK8 A0A0L7RFR4 A0A3L8E056 A0A0M8ZTR8 A0A026WY78 A0A1B6EEU7 A0A1B6C766 A0A154PBN6 A0A067QJ98 A0A195D392 A0A151IUV5 A0A195FIM5 F4WUT5 A0A182IXV5 A0A158P2E7 A0A151WUA5 A0A182V600 A0A232EJ91 A0A182M7Y0 A0A3F2Z265 A0A182LKK2 A0A182I4B9 Q7Q5B2 A0A084WMM0 A0A1B6K9V5 A0A0L0CRF4 A0A2M4AS70 A0A2M4BLH3 B0W850 E9IGD4 A0A1Q3FDH8 A0A2M4BL57 Q16QM2 A0A182GFB1 A0A2M4CTW7 T1PAR7 W8BHI9 A0A1B6IHH1 A0A182NIV9 A0A195BTG4 A0A0A1XMV6 A0A310SE40 A0A182FFE3 A0A182QE10 A0A2M4CTX6 A0A1I8PLC7 A0A0K8W820 A0A1W4UX18 B4HUZ8 B3NFK6 B4QJV3 O02373 A0A0M4EZF1 B4LF30 B4PJT5 B4J387 A0A2J7QSV3 B3M9F6 B4MKT9 A0A1Y1N4K7 A0A1B0D3J1 A0A1A9X520 A0A3B0KN01 A0A1L8E1X7 A0A1B0GJL9 A0A023ESW2 W8H0S7 A0A1W4WEY7 B4KZU4 Q29F37 A0A1A9UXX4 A0A1B0B9F7 A0A1A9Z3J4 A0A1A9Y9H5 A0A1W4WPX2 A0A0C9RD46 A0A0C9RM96 A0A3Q0ILJ3 T1IY28 J3JZE6 A0A146LDX8 E0VZ63 A0A182WBV8 A0A1B0FC52 A0A1J1HZZ0 A0A2A3EB85
A0A212EYQ8 A0A0L7L164 A0A069DTZ0 A0A023F4G4 T1HCB1 K7J7N2 E2AU70 A0A088A2I2 A0A0P4VSK8 A0A0L7RFR4 A0A3L8E056 A0A0M8ZTR8 A0A026WY78 A0A1B6EEU7 A0A1B6C766 A0A154PBN6 A0A067QJ98 A0A195D392 A0A151IUV5 A0A195FIM5 F4WUT5 A0A182IXV5 A0A158P2E7 A0A151WUA5 A0A182V600 A0A232EJ91 A0A182M7Y0 A0A3F2Z265 A0A182LKK2 A0A182I4B9 Q7Q5B2 A0A084WMM0 A0A1B6K9V5 A0A0L0CRF4 A0A2M4AS70 A0A2M4BLH3 B0W850 E9IGD4 A0A1Q3FDH8 A0A2M4BL57 Q16QM2 A0A182GFB1 A0A2M4CTW7 T1PAR7 W8BHI9 A0A1B6IHH1 A0A182NIV9 A0A195BTG4 A0A0A1XMV6 A0A310SE40 A0A182FFE3 A0A182QE10 A0A2M4CTX6 A0A1I8PLC7 A0A0K8W820 A0A1W4UX18 B4HUZ8 B3NFK6 B4QJV3 O02373 A0A0M4EZF1 B4LF30 B4PJT5 B4J387 A0A2J7QSV3 B3M9F6 B4MKT9 A0A1Y1N4K7 A0A1B0D3J1 A0A1A9X520 A0A3B0KN01 A0A1L8E1X7 A0A1B0GJL9 A0A023ESW2 W8H0S7 A0A1W4WEY7 B4KZU4 Q29F37 A0A1A9UXX4 A0A1B0B9F7 A0A1A9Z3J4 A0A1A9Y9H5 A0A1W4WPX2 A0A0C9RD46 A0A0C9RM96 A0A3Q0ILJ3 T1IY28 J3JZE6 A0A146LDX8 E0VZ63 A0A182WBV8 A0A1B0FC52 A0A1J1HZZ0 A0A2A3EB85
EC Number
1.1.1.22
Pubmed
19121390
26354079
28756777
22118469
26227816
26334808
+ More
25474469 20075255 20798317 27129103 30249741 24508170 24845553 21719571 21347285 28648823 20966253 12364791 14747013 17210077 24438588 26108605 21282665 17510324 26483478 25315136 24495485 25830018 17994087 22936249 9217004 9342049 9272947 10731132 12537572 12537569 17550304 28004739 24945155 15632085 22516182 26823975 20566863
25474469 20075255 20798317 27129103 30249741 24508170 24845553 21719571 21347285 28648823 20966253 12364791 14747013 17210077 24438588 26108605 21282665 17510324 26483478 25315136 24495485 25830018 17994087 22936249 9217004 9342049 9272947 10731132 12537572 12537569 17550304 28004739 24945155 15632085 22516182 26823975 20566863
EMBL
BABH01031771
BABH01031772
BABH01031773
KQ458863
KPJ04714.1
KQ459988
+ More
KPJ18783.1 NWSH01002937 PCG67251.1 ODYU01001796 SOQ38488.1 KZ150412 PZC70975.1 AGBW02011451 OWR46628.1 JTDY01003777 KOB69019.1 GBGD01001316 JAC87573.1 GBBI01002292 JAC16420.1 ACPB03000774 AAZX01012151 GL442807 EFN63018.1 GDKW01001144 JAI55451.1 KQ414601 KOC69675.1 QOIP01000002 RLU26084.1 KQ435881 KOX69806.1 KK107069 EZA60783.1 GEDC01026031 GEDC01000868 JAS11267.1 JAS36430.1 GEDC01028034 GEDC01007246 JAS09264.1 JAS30052.1 KQ434869 KZC09261.1 KK853297 KDR08787.1 KQ976885 KYN07380.1 KQ980945 KYN11251.1 KQ981523 KYN40221.1 GL888375 EGI62082.1 ADTU01007246 ADTU01007247 KQ982753 KYQ51215.1 NNAY01004096 OXU18371.1 AXCM01000272 AXCM01000273 APCN01002300 APCN01002301 AAAB01008960 EAA11440.4 ATLV01024482 KE525352 KFB51464.1 GEBQ01031751 JAT08226.1 JRES01000122 KNC34014.1 GGFK01010117 MBW43438.1 GGFJ01004670 MBW53811.1 DS231857 EDS38675.1 GL762997 EFZ20353.1 GFDL01009493 JAV25552.1 GGFJ01004669 MBW53810.1 CH477744 EAT36695.1 JXUM01000833 JXUM01000834 JXUM01000835 JXUM01000836 GGFL01004527 MBW68705.1 KA645799 AFP60428.1 GAMC01008343 JAB98212.1 GECU01021334 GECU01005295 JAS86372.1 JAT02412.1 KQ976408 KYM91226.1 GBXI01001981 JAD12311.1 KQ771508 OAD52535.1 AXCN02000476 GGFL01004551 MBW68729.1 GDHF01005289 JAI47025.1 CH480817 EDW50769.1 CH954178 EDV50548.1 CM000363 CM002912 EDX09494.1 KMY97976.1 AF007870 AF000570 AF009013 AF001310 AH007072 AE014296 AY052137 CP012525 ALC43757.1 CH940647 EDW69199.1 CM000159 EDW93684.1 CH916366 EDV96158.1 NEVH01011219 PNF31655.1 CH902618 EDV41169.1 CH963847 EDW72795.1 GEZM01013056 JAV92794.1 AJVK01023578 AJVK01023579 OUUW01000009 SPP85188.1 GFDF01001344 JAV12740.1 AJWK01021979 GAPW01001208 JAC12390.1 KF986615 AHK26793.1 CH933809 EDW17956.1 CH379068 EAL31235.1 JXJN01010371 GBYB01014514 JAG84281.1 GBYB01014512 JAG84279.1 JH431669 BT128627 AEE63584.1 GDHC01012201 JAQ06428.1 DS235849 EEB18669.1 CCAG010016068 CVRI01000035 CRK92884.1 KZ288311 PBC28446.1
KPJ18783.1 NWSH01002937 PCG67251.1 ODYU01001796 SOQ38488.1 KZ150412 PZC70975.1 AGBW02011451 OWR46628.1 JTDY01003777 KOB69019.1 GBGD01001316 JAC87573.1 GBBI01002292 JAC16420.1 ACPB03000774 AAZX01012151 GL442807 EFN63018.1 GDKW01001144 JAI55451.1 KQ414601 KOC69675.1 QOIP01000002 RLU26084.1 KQ435881 KOX69806.1 KK107069 EZA60783.1 GEDC01026031 GEDC01000868 JAS11267.1 JAS36430.1 GEDC01028034 GEDC01007246 JAS09264.1 JAS30052.1 KQ434869 KZC09261.1 KK853297 KDR08787.1 KQ976885 KYN07380.1 KQ980945 KYN11251.1 KQ981523 KYN40221.1 GL888375 EGI62082.1 ADTU01007246 ADTU01007247 KQ982753 KYQ51215.1 NNAY01004096 OXU18371.1 AXCM01000272 AXCM01000273 APCN01002300 APCN01002301 AAAB01008960 EAA11440.4 ATLV01024482 KE525352 KFB51464.1 GEBQ01031751 JAT08226.1 JRES01000122 KNC34014.1 GGFK01010117 MBW43438.1 GGFJ01004670 MBW53811.1 DS231857 EDS38675.1 GL762997 EFZ20353.1 GFDL01009493 JAV25552.1 GGFJ01004669 MBW53810.1 CH477744 EAT36695.1 JXUM01000833 JXUM01000834 JXUM01000835 JXUM01000836 GGFL01004527 MBW68705.1 KA645799 AFP60428.1 GAMC01008343 JAB98212.1 GECU01021334 GECU01005295 JAS86372.1 JAT02412.1 KQ976408 KYM91226.1 GBXI01001981 JAD12311.1 KQ771508 OAD52535.1 AXCN02000476 GGFL01004551 MBW68729.1 GDHF01005289 JAI47025.1 CH480817 EDW50769.1 CH954178 EDV50548.1 CM000363 CM002912 EDX09494.1 KMY97976.1 AF007870 AF000570 AF009013 AF001310 AH007072 AE014296 AY052137 CP012525 ALC43757.1 CH940647 EDW69199.1 CM000159 EDW93684.1 CH916366 EDV96158.1 NEVH01011219 PNF31655.1 CH902618 EDV41169.1 CH963847 EDW72795.1 GEZM01013056 JAV92794.1 AJVK01023578 AJVK01023579 OUUW01000009 SPP85188.1 GFDF01001344 JAV12740.1 AJWK01021979 GAPW01001208 JAC12390.1 KF986615 AHK26793.1 CH933809 EDW17956.1 CH379068 EAL31235.1 JXJN01010371 GBYB01014514 JAG84281.1 GBYB01014512 JAG84279.1 JH431669 BT128627 AEE63584.1 GDHC01012201 JAQ06428.1 DS235849 EEB18669.1 CCAG010016068 CVRI01000035 CRK92884.1 KZ288311 PBC28446.1
Proteomes
UP000005204
UP000053268
UP000053240
UP000218220
UP000007151
UP000037510
+ More
UP000015103 UP000002358 UP000000311 UP000005203 UP000053825 UP000279307 UP000053105 UP000053097 UP000076502 UP000027135 UP000078542 UP000078492 UP000078541 UP000007755 UP000075880 UP000005205 UP000075809 UP000075903 UP000215335 UP000075883 UP000076407 UP000075882 UP000075840 UP000007062 UP000030765 UP000037069 UP000002320 UP000008820 UP000069940 UP000095301 UP000075884 UP000078540 UP000069272 UP000075886 UP000095300 UP000192221 UP000001292 UP000008711 UP000000304 UP000000803 UP000092553 UP000008792 UP000002282 UP000001070 UP000235965 UP000007801 UP000007798 UP000092462 UP000091820 UP000268350 UP000092461 UP000192223 UP000009192 UP000001819 UP000078200 UP000092460 UP000092445 UP000092443 UP000079169 UP000009046 UP000075920 UP000092444 UP000183832 UP000242457
UP000015103 UP000002358 UP000000311 UP000005203 UP000053825 UP000279307 UP000053105 UP000053097 UP000076502 UP000027135 UP000078542 UP000078492 UP000078541 UP000007755 UP000075880 UP000005205 UP000075809 UP000075903 UP000215335 UP000075883 UP000076407 UP000075882 UP000075840 UP000007062 UP000030765 UP000037069 UP000002320 UP000008820 UP000069940 UP000095301 UP000075884 UP000078540 UP000069272 UP000075886 UP000095300 UP000192221 UP000001292 UP000008711 UP000000304 UP000000803 UP000092553 UP000008792 UP000002282 UP000001070 UP000235965 UP000007801 UP000007798 UP000092462 UP000091820 UP000268350 UP000092461 UP000192223 UP000009192 UP000001819 UP000078200 UP000092460 UP000092445 UP000092443 UP000079169 UP000009046 UP000075920 UP000092444 UP000183832 UP000242457
Interpro
IPR028356
UDPglc_DH_euk
+ More
IPR014027 UDP-Glc/GDP-Man_DH_C
IPR017476 UDP-Glc/GDP-Man
IPR036220 UDP-Glc/GDP-Man_DH_C_sf
IPR014026 UDP-Glc/GDP-Man_DH_dimer
IPR008927 6-PGluconate_DH-like_C_sf
IPR036291 NAD(P)-bd_dom_sf
IPR001732 UDP-Glc/GDP-Man_DH_N
IPR002994 Surf1/Shy1
IPR013087 Znf_C2H2_type
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR001680 WD40_repeat
IPR017986 WD40_repeat_dom
IPR036322 WD40_repeat_dom_sf
IPR014027 UDP-Glc/GDP-Man_DH_C
IPR017476 UDP-Glc/GDP-Man
IPR036220 UDP-Glc/GDP-Man_DH_C_sf
IPR014026 UDP-Glc/GDP-Man_DH_dimer
IPR008927 6-PGluconate_DH-like_C_sf
IPR036291 NAD(P)-bd_dom_sf
IPR001732 UDP-Glc/GDP-Man_DH_N
IPR002994 Surf1/Shy1
IPR013087 Znf_C2H2_type
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR001680 WD40_repeat
IPR017986 WD40_repeat_dom
IPR036322 WD40_repeat_dom_sf
Gene 3D
CDD
ProteinModelPortal
H9JRM8
A0A0N1I5K8
A0A0N1IQ45
A0A2A4J5K4
A0A2H1VCB3
A0A2W1B998
+ More
A0A212EYQ8 A0A0L7L164 A0A069DTZ0 A0A023F4G4 T1HCB1 K7J7N2 E2AU70 A0A088A2I2 A0A0P4VSK8 A0A0L7RFR4 A0A3L8E056 A0A0M8ZTR8 A0A026WY78 A0A1B6EEU7 A0A1B6C766 A0A154PBN6 A0A067QJ98 A0A195D392 A0A151IUV5 A0A195FIM5 F4WUT5 A0A182IXV5 A0A158P2E7 A0A151WUA5 A0A182V600 A0A232EJ91 A0A182M7Y0 A0A3F2Z265 A0A182LKK2 A0A182I4B9 Q7Q5B2 A0A084WMM0 A0A1B6K9V5 A0A0L0CRF4 A0A2M4AS70 A0A2M4BLH3 B0W850 E9IGD4 A0A1Q3FDH8 A0A2M4BL57 Q16QM2 A0A182GFB1 A0A2M4CTW7 T1PAR7 W8BHI9 A0A1B6IHH1 A0A182NIV9 A0A195BTG4 A0A0A1XMV6 A0A310SE40 A0A182FFE3 A0A182QE10 A0A2M4CTX6 A0A1I8PLC7 A0A0K8W820 A0A1W4UX18 B4HUZ8 B3NFK6 B4QJV3 O02373 A0A0M4EZF1 B4LF30 B4PJT5 B4J387 A0A2J7QSV3 B3M9F6 B4MKT9 A0A1Y1N4K7 A0A1B0D3J1 A0A1A9X520 A0A3B0KN01 A0A1L8E1X7 A0A1B0GJL9 A0A023ESW2 W8H0S7 A0A1W4WEY7 B4KZU4 Q29F37 A0A1A9UXX4 A0A1B0B9F7 A0A1A9Z3J4 A0A1A9Y9H5 A0A1W4WPX2 A0A0C9RD46 A0A0C9RM96 A0A3Q0ILJ3 T1IY28 J3JZE6 A0A146LDX8 E0VZ63 A0A182WBV8 A0A1B0FC52 A0A1J1HZZ0 A0A2A3EB85
A0A212EYQ8 A0A0L7L164 A0A069DTZ0 A0A023F4G4 T1HCB1 K7J7N2 E2AU70 A0A088A2I2 A0A0P4VSK8 A0A0L7RFR4 A0A3L8E056 A0A0M8ZTR8 A0A026WY78 A0A1B6EEU7 A0A1B6C766 A0A154PBN6 A0A067QJ98 A0A195D392 A0A151IUV5 A0A195FIM5 F4WUT5 A0A182IXV5 A0A158P2E7 A0A151WUA5 A0A182V600 A0A232EJ91 A0A182M7Y0 A0A3F2Z265 A0A182LKK2 A0A182I4B9 Q7Q5B2 A0A084WMM0 A0A1B6K9V5 A0A0L0CRF4 A0A2M4AS70 A0A2M4BLH3 B0W850 E9IGD4 A0A1Q3FDH8 A0A2M4BL57 Q16QM2 A0A182GFB1 A0A2M4CTW7 T1PAR7 W8BHI9 A0A1B6IHH1 A0A182NIV9 A0A195BTG4 A0A0A1XMV6 A0A310SE40 A0A182FFE3 A0A182QE10 A0A2M4CTX6 A0A1I8PLC7 A0A0K8W820 A0A1W4UX18 B4HUZ8 B3NFK6 B4QJV3 O02373 A0A0M4EZF1 B4LF30 B4PJT5 B4J387 A0A2J7QSV3 B3M9F6 B4MKT9 A0A1Y1N4K7 A0A1B0D3J1 A0A1A9X520 A0A3B0KN01 A0A1L8E1X7 A0A1B0GJL9 A0A023ESW2 W8H0S7 A0A1W4WEY7 B4KZU4 Q29F37 A0A1A9UXX4 A0A1B0B9F7 A0A1A9Z3J4 A0A1A9Y9H5 A0A1W4WPX2 A0A0C9RD46 A0A0C9RM96 A0A3Q0ILJ3 T1IY28 J3JZE6 A0A146LDX8 E0VZ63 A0A182WBV8 A0A1B0FC52 A0A1J1HZZ0 A0A2A3EB85
PDB
5W4X
E-value=1.58623e-167,
Score=1513
Ontologies
KEGG
PATHWAY
GO
PANTHER
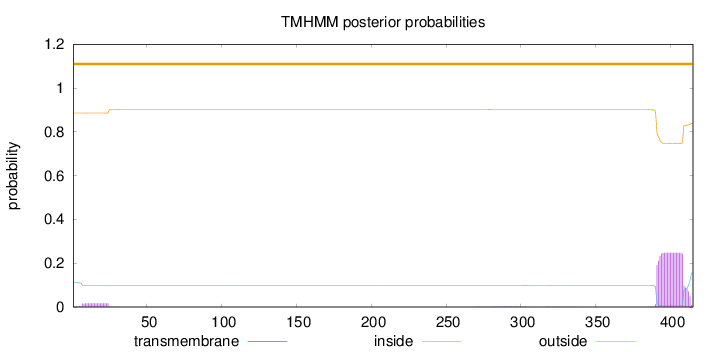
Topology
Length:
415
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
5.10674
Exp number, first 60 AAs:
0.31921
Total prob of N-in:
0.11371
outside
1 - 415
Population Genetic Test Statistics
Pi
222.702548
Theta
189.30514
Tajima's D
0.59448
CLR
0.307851
CSRT
0.543172841357932
Interpretation
Uncertain
