Gene
KWMTBOMO14867
Annotation
endonuclease-reverse_transcriptase_[Bombyx_mori]
Full name
Sodium/hydrogen exchanger
Location in the cell
Nuclear Reliability : 3.608
Sequence
CDS
ATGGAATTACTTCTACGTAAACTCGAAGAGAAATTAGATAAACAAGCCGACCAGATAACAAAGGCGGTAACTAAGAACGTCACAGAAGCTATAGACGAGAAAATAAATGCGATAATTGAAGAAAATAAACTCCTCACAAACAAAGTATCCGAACTAGAGTTGAAAATTAAATCCTTAGAAAGAGAGAAACGAAAAAACAACCTCGTCTTCTTCGGAGTAGACGAAATAGGAAAATCCGAGCGAGAATTAGTAGATTATATAAAAGATACCATAGAAGATCTAGGAGTGCAAATGAACAGCCAAGAGATTAGTAACATATATAGAATCGGAAGAAAGACGGAAAACAAAAACCGACCAGTCGTAATTTCTTTAACAACCCTGTGGAAGAAGCATCTTGTACTCAAGAACAAGTCTAATCTCCCGTCAAATATATATGTTAAAGAGGATTACTCCAAGGAAACTCTGGAAAGGCGCAAAAAACTACAATCCCAAGTGGAAGAAGAAAAAAAGAAAGAAAACCGGAAACAAATAGAAACGGCTAAAACAATTAGACTTAAGCAAAAACAACGAACAAATTGTCTCCCACAACGGCTGGTCTCCGCGGGAGCACTAGACCAAAACCCTCCAGCAAAAAGAAACGAAACATTCCACATTGCAACACTCAACACAAGAACTCTGCGAACACAGGAAAGCCTACTTGAACTGGACAAGGCGTTACAAAATATAAAACTGGATATCCTGGGAATATGTGAAATGAGAAGGCTTGATGAAAAAATAACTGAACGAAACAACTACATATTATATCAGAAAGGTGAAACAGCAGGCCAAAGAGGTGTGGGCTTCTTAATTAAAAAATCATTGAAGAACTTTATACAAGAACTGATAGGCATTTCAGACAGAATAGCAATACTCAACATTAACTTACCAAGATACAAAAAACCCTGGACAATTATACAAGCATACGCACCCACAGAATTAGCATTGGAAAGTGAAAGAGAGGGATTTTACTGCACACTATCTGATATTATAAAAAAATATTATAAGAACTTTCTCATAGTCATGGGTGACTTTAATGCACAAGTTGGCTGTAGAGAACCCGGAGAAGAACTGATAATGGGCAAACATGGATTTGGAAAGAGGAGTGCAAACGGACATCGACTAATTGAATTTTTACTACAAAACAACTTAACAATACTAAATTCGCTCTACAAAAAAAAACCCAAAACCAAGTGGACTTGGATATCACCTGATGGAAAATATAAGAATGAAATCGATTTCATAATAACTAACAAACCAAAGTTTTTCCAAGATACACACATTATTCAAAACTTAAATTTTAATACGAACCACCGTATGGTACGTTGCCGTTTAGAAAAGCATCAACCAAAAAGAAGTAGAAATAATATTAAAAATAAATTGACATTAGAACAACACAAAGAAAAAATAGATTCATCGTCCATCAACACACTGAAAGAAATAGTAAACTCGAACAGAAGTGTCACAGAAAAATATGATCTCATGGTCACACAACTGTCAAAATTGAGGAAAGTAAATGCAACAAAAGATGTGAAGTTTGGTGCGAAGACAATGGAGCTCCTAGAAGAACGAAAAGTACTTATAACAGACAAATCAAAAACTGAAGAAAACCGTAAGTTAATATCTAAACTGAGCAAAGAGATTCGGGAAAGCATAAGAAAGGATAGGAAATTAAGAAGAACGAAAAAATTAGAAGATGAAATTCTAAGAACAGGGGGAACAAGAAAAGCTTTTAAGGAATTAAGAGAACATGGAGTGGAATGGATACCAAAACTAAAAAGCAACAGCTCCAAACAAGCAACAAATCGCAGAAAAATTCAGAGTCTTGCTACAGATTACTATCGAACTCTTTACTCGGACAATGACACCGACCGAGAAAAGACCGCCGTTAACGATCCTAGTACGAATAATAAAAAAGTACCACCAGTACTCTTGCAAGAAGTACAGAAAGCCATCATGTCTCAGAAGCCAGAAAAATCTCCAGGACCCGACAGTATATCAAATGAGCTGATTAGAGGTAACGAAGAAGAAATAAGCCCAATCCTAACCAAGCTATTCAATGATATTGTAACAACAGGTGTCATCCCAAAACAATGGAATGAGTCACATATCATCCTCCTCCATAAGAAAGGAGATAAAGATGATATTGGCAACTACAGACCAATCAGTCTAATATCAAATGTGTACAAAGTCTTCGCAAAAGTAATTCTAGAAAGAATAACAAGGGCTTTAGACGAAAACCAACCGATCGAGCAGGCAGGATTTAGAAAGAACTACAGCACTATAGACCACATCCACACAATCAAACAAGTTTTGGAAAAATACAATGAATACCAGAAGACTTTGTATATCTGCTTTATAGACTACTCCAAAGCATTTGACAGTATAAAACACAAATTTATTTGGCTGAGTCTAAAACAACAAGGAGTGGACGATATATATATAGAACTAATCAAATCCATCTACAAAAAAAGTAAAAGCAGTATTAAATTAGAGTCAAAAGGAGACCCCTTTCCAATAAACAGAGGCGTAAGACAAGGTGACCCATTATCTCCCAAGCTGTTTAATGCAGTACTGGAATGCACATTTAGAAGATTAAACTGGGATAACTATGGTATTATAATCAATGGAAGTCGCCTCAATCATTTGAGATTTGCAGATGATATTATCTTGCTAGAAGAAGATCCTAAGAAACTTGAACACATGATTAAAGAAATAGTAGAAAAAAGCAGAGAAGTAGGGCTAGAAATTAATGCAGATAAAACGAAACTAATGACGAACTCAACAGAAAGGAACATAGAGGTAAACGGAACCAAGTTCGAATATGTGAAGGAATTCGTTTACCTAGGACAAATTATATCTTCTGAAGACTCCATGTCCAAGGAAATAAACAAAAGAATAGCATGCGGTTGGAAAAAATACTGGTCCTTGAAAGAGATAATGAAGAATAAAGATCTAGGTATACATATCAAGAGAAAGACTTTCAACACTTGTATTCTCCCTTGCATGACTTACGGATGTGAGACATGGACACTAACTGACAAACATAGAGAAAAACTTGCTAAAACAGAGAGAGCAATGGAAAGAAGCATGTTGAGTATCAGACTTAGTGACAGGATTCGAAATTGGGAAATAAGAAGGAAGACAAAAGTAACGGATATAATAACTCGTATAGAAAATCTTAAATGGAGTTGGACAGGACATATGCTACGTTGCAAAAAAAACAAGTGGAGCAAACAAGTTACTTTATGGTATCCAAGAGGGGACACCAGAAGACAGGGGCGACCAAAAACGAGATGGAGCGATGATATACGCCTAACCCTAGGACCATACTGGACCAGAGTTGCAGAGGACCGAGCCCAATGGAGAGAGCTGGAGGAGGCCTATGCCACGAGGCACGCCGAACTCCGAGATATACTATAA
Protein
MELLLRKLEEKLDKQADQITKAVTKNVTEAIDEKINAIIEENKLLTNKVSELELKIKSLEREKRKNNLVFFGVDEIGKSERELVDYIKDTIEDLGVQMNSQEISNIYRIGRKTENKNRPVVISLTTLWKKHLVLKNKSNLPSNIYVKEDYSKETLERRKKLQSQVEEEKKKENRKQIETAKTIRLKQKQRTNCLPQRLVSAGALDQNPPAKRNETFHIATLNTRTLRTQESLLELDKALQNIKLDILGICEMRRLDEKITERNNYILYQKGETAGQRGVGFLIKKSLKNFIQELIGISDRIAILNINLPRYKKPWTIIQAYAPTELALESEREGFYCTLSDIIKKYYKNFLIVMGDFNAQVGCREPGEELIMGKHGFGKRSANGHRLIEFLLQNNLTILNSLYKKKPKTKWTWISPDGKYKNEIDFIITNKPKFFQDTHIIQNLNFNTNHRMVRCRLEKHQPKRSRNNIKNKLTLEQHKEKIDSSSINTLKEIVNSNRSVTEKYDLMVTQLSKLRKVNATKDVKFGAKTMELLEERKVLITDKSKTEENRKLISKLSKEIRESIRKDRKLRRTKKLEDEILRTGGTRKAFKELREHGVEWIPKLKSNSSKQATNRRKIQSLATDYYRTLYSDNDTDREKTAVNDPSTNNKKVPPVLLQEVQKAIMSQKPEKSPGPDSISNELIRGNEEEISPILTKLFNDIVTTGVIPKQWNESHIILLHKKGDKDDIGNYRPISLISNVYKVFAKVILERITRALDENQPIEQAGFRKNYSTIDHIHTIKQVLEKYNEYQKTLYICFIDYSKAFDSIKHKFIWLSLKQQGVDDIYIELIKSIYKKSKSSIKLESKGDPFPINRGVRQGDPLSPKLFNAVLECTFRRLNWDNYGIIINGSRLNHLRFADDIILLEEDPKKLEHMIKEIVEKSREVGLEINADKTKLMTNSTERNIEVNGTKFEYVKEFVYLGQIISSEDSMSKEINKRIACGWKKYWSLKEIMKNKDLGIHIKRKTFNTCILPCMTYGCETWTLTDKHREKLAKTERAMERSMLSIRLSDRIRNWEIRRKTKVTDIITRIENLKWSWTGHMLRCKKNKWSKQVTLWYPRGDTRRQGRPKTRWSDDIRLTLGPYWTRVAEDRAQWRELEEAYATRHAELRDIL
Summary
Similarity
Belongs to the monovalent cation:proton antiporter 1 (CPA1) transporter (TC 2.A.36) family.
Feature
chain Sodium/hydrogen exchanger
Uniprot
D7F179
D7F176
A0A2H1WF27
D7F177
D7F178
A0A023EXV2
+ More
A0A2W1BZV1 A0A2H1VS24 A0A069DZM5 D7F157 D7F164 D7F165 D7F166 D7F160 A0A016S1D2 A0A016TK11 A0A016SG46 A0A016W4R1 E3NC15 E3MH09 A0A016WPY4 A0A016WCB8 A0A016ULU9 A0A016VEC1 A0A016VCD9 A0A016T590 A0A016T7J7 A0A016VCU3 A0A016VBP7 A0A0K0FAW8 A0A016V6A2 E3NGJ2 A0A016S5B2 E3N0F4 A0A016VVN3 A0A016S795 A0A016WA79 A0A016UN79 Q9TZX4 A0A016SX67 K7IG35 A0A2H2J8I4 A0A016V419 G0MZ46 A0A2H2JFN0 K7ICT2 K7ICT1 A0A016RUR3 A0A016RV73 K7H0R0 K7IBZ6 K7H0Q9 K7I2A3 K7IBZ5 A0A016SLV8 K7IG36 K7HDM2 K7HDM3 A0A016RRX8 E3NJQ7 A0A016THC6 A0A016U352 A0A016U3L3 A0A016T4C4 A0A016SGC3 A0A016SVA2 K7I2A2 A0A016SW37 A0A016W1X2 A0A016WYN9 A0A016SPR9 A0A016SCZ8 K7H0Q8 A0A016SCL7 A0A016VLP0 A0A016SY13 A0A016U6B7 A0A016VCK8 A0A016U123 K7I2A1 A0A016X207 A0A016WFS9 A0A016SCT9 A0A016VT25 A0A2W5U8K4 A0A016UJS4 A0A016WE57 A0A016T9D3 A0A016TK92 A0A016TQJ1 A0A016T9A5 A0A016VGA0 A0A016WUN2
A0A2W1BZV1 A0A2H1VS24 A0A069DZM5 D7F157 D7F164 D7F165 D7F166 D7F160 A0A016S1D2 A0A016TK11 A0A016SG46 A0A016W4R1 E3NC15 E3MH09 A0A016WPY4 A0A016WCB8 A0A016ULU9 A0A016VEC1 A0A016VCD9 A0A016T590 A0A016T7J7 A0A016VCU3 A0A016VBP7 A0A0K0FAW8 A0A016V6A2 E3NGJ2 A0A016S5B2 E3N0F4 A0A016VVN3 A0A016S795 A0A016WA79 A0A016UN79 Q9TZX4 A0A016SX67 K7IG35 A0A2H2J8I4 A0A016V419 G0MZ46 A0A2H2JFN0 K7ICT2 K7ICT1 A0A016RUR3 A0A016RV73 K7H0R0 K7IBZ6 K7H0Q9 K7I2A3 K7IBZ5 A0A016SLV8 K7IG36 K7HDM2 K7HDM3 A0A016RRX8 E3NJQ7 A0A016THC6 A0A016U352 A0A016U3L3 A0A016T4C4 A0A016SGC3 A0A016SVA2 K7I2A2 A0A016SW37 A0A016W1X2 A0A016WYN9 A0A016SPR9 A0A016SCZ8 K7H0Q8 A0A016SCL7 A0A016VLP0 A0A016SY13 A0A016U6B7 A0A016VCK8 A0A016U123 K7I2A1 A0A016X207 A0A016WFS9 A0A016SCT9 A0A016VT25 A0A2W5U8K4 A0A016UJS4 A0A016WE57 A0A016T9D3 A0A016TK92 A0A016TQJ1 A0A016T9A5 A0A016VGA0 A0A016WUN2
EMBL
FJ265564
ADI61832.1
FJ265561
ADI61829.1
ODYU01008249
SOQ51673.1
+ More
FJ265562 ADI61830.1 FJ265563 ADI61831.1 GBBI01004876 JAC13836.1 KZ149907 PZC78256.1 ODYU01004088 SOQ43598.1 GBGD01000380 JAC88509.1 FJ265542 ADI61810.1 FJ265549 ADI61817.1 FJ265550 ADI61818.1 FJ265551 ADI61819.1 FJ265545 ADI61813.1 JARK01001655 EYB84321.1 JARK01001676 JARK01001446 JARK01001433 EYB83199.1 EYC01126.1 EYC02928.1 JARK01001565 EYB89683.1 JARK01001337 EYC34625.1 DS268591 EFO92565.1 DS268444 EFP01729.1 JARK01000173 EYC41342.1 JARK01000388 EYC37469.1 JARK01001370 EYC16150.1 JARK01001348 EYC25048.1 EYC25050.1 JARK01001473 EYB97806.1 JARK01001736 JARK01001462 JARK01001379 JARK01001338 EYB80775.1 EYB98943.1 EYC13646.1 EYC33347.1 EYC25046.1 EYC25049.1 JARK01001353 EYC22263.1 DS268655 EFO97114.1 JARK01001626 EYB85833.1 DS268505 EFP13372.1 JARK01001340 EYC30828.1 JARK01001621 EYB86109.1 JARK01000460 EYC36749.1 EYC16376.1 AF054983 AAC72298.1 JARK01001500 JARK01001343 EYB95040.1 EYC28825.1 JARK01001354 EYC21986.1 GL379822 EGT48339.1 JARK01001711 EYB81734.1 JARK01001700 JARK01001393 EYB82228.1 EYC10181.1 JARK01001543 EYB91356.1 JARK01001730 EYB81048.1 DS268752 EFP01106.1 JARK01001438 EYC02111.1 JARK01001395 EYC09729.1 EYC09730.1 JARK01001475 EYB97550.1 JARK01001568 EYB89432.1 JARK01001505 EYB94618.1 EYB94617.1 EYC33297.1 JARK01000063 EYC44387.1 JARK01001531 EYB92324.1 JARK01001581 EYB88505.1 JARK01001628 JARK01001583 JARK01001528 JARK01001506 JARK01001470 JARK01001467 JARK01001463 JARK01001457 JARK01001432 JARK01001420 JARK01001403 JARK01001371 JARK01001369 JARK01001360 JARK01001349 JARK01000603 JARK01000080 EYB85741.1 EYB88438.1 EYB92544.1 EYB94540.1 EYB98080.1 EYB98468.1 EYB98899.1 EYB99568.1 EYC03088.1 EYC05014.1 EYC08153.1 EYC15852.1 EYC16408.1 EYC19456.1 EYC22445.1 EYC24734.1 EYC24778.1 EYC24866.1 EYC33488.1 EYC35672.1 EYC43798.1 JARK01001344 EYC28216.1 JARK01001497 EYB95322.1 JARK01001389 EYC10874.1 EYC25145.1 JARK01001401 EYC08686.1 JARK01000008 EYC46079.1 JARK01000302 EYC38674.1 JARK01001582 EYB88498.1 EYC30744.1 QFQQ01000179 PZR20596.1 JARK01001372 EYC15614.1 JARK01000343 EYC38099.1 JARK01001459 EYB99276.1 EYC03125.1 JARK01001421 EYC04912.1 EYB99275.1 JARK01001346 EYC26445.1 JARK01000109 EYC42952.1
FJ265562 ADI61830.1 FJ265563 ADI61831.1 GBBI01004876 JAC13836.1 KZ149907 PZC78256.1 ODYU01004088 SOQ43598.1 GBGD01000380 JAC88509.1 FJ265542 ADI61810.1 FJ265549 ADI61817.1 FJ265550 ADI61818.1 FJ265551 ADI61819.1 FJ265545 ADI61813.1 JARK01001655 EYB84321.1 JARK01001676 JARK01001446 JARK01001433 EYB83199.1 EYC01126.1 EYC02928.1 JARK01001565 EYB89683.1 JARK01001337 EYC34625.1 DS268591 EFO92565.1 DS268444 EFP01729.1 JARK01000173 EYC41342.1 JARK01000388 EYC37469.1 JARK01001370 EYC16150.1 JARK01001348 EYC25048.1 EYC25050.1 JARK01001473 EYB97806.1 JARK01001736 JARK01001462 JARK01001379 JARK01001338 EYB80775.1 EYB98943.1 EYC13646.1 EYC33347.1 EYC25046.1 EYC25049.1 JARK01001353 EYC22263.1 DS268655 EFO97114.1 JARK01001626 EYB85833.1 DS268505 EFP13372.1 JARK01001340 EYC30828.1 JARK01001621 EYB86109.1 JARK01000460 EYC36749.1 EYC16376.1 AF054983 AAC72298.1 JARK01001500 JARK01001343 EYB95040.1 EYC28825.1 JARK01001354 EYC21986.1 GL379822 EGT48339.1 JARK01001711 EYB81734.1 JARK01001700 JARK01001393 EYB82228.1 EYC10181.1 JARK01001543 EYB91356.1 JARK01001730 EYB81048.1 DS268752 EFP01106.1 JARK01001438 EYC02111.1 JARK01001395 EYC09729.1 EYC09730.1 JARK01001475 EYB97550.1 JARK01001568 EYB89432.1 JARK01001505 EYB94618.1 EYB94617.1 EYC33297.1 JARK01000063 EYC44387.1 JARK01001531 EYB92324.1 JARK01001581 EYB88505.1 JARK01001628 JARK01001583 JARK01001528 JARK01001506 JARK01001470 JARK01001467 JARK01001463 JARK01001457 JARK01001432 JARK01001420 JARK01001403 JARK01001371 JARK01001369 JARK01001360 JARK01001349 JARK01000603 JARK01000080 EYB85741.1 EYB88438.1 EYB92544.1 EYB94540.1 EYB98080.1 EYB98468.1 EYB98899.1 EYB99568.1 EYC03088.1 EYC05014.1 EYC08153.1 EYC15852.1 EYC16408.1 EYC19456.1 EYC22445.1 EYC24734.1 EYC24778.1 EYC24866.1 EYC33488.1 EYC35672.1 EYC43798.1 JARK01001344 EYC28216.1 JARK01001497 EYB95322.1 JARK01001389 EYC10874.1 EYC25145.1 JARK01001401 EYC08686.1 JARK01000008 EYC46079.1 JARK01000302 EYC38674.1 JARK01001582 EYB88498.1 EYC30744.1 QFQQ01000179 PZR20596.1 JARK01001372 EYC15614.1 JARK01000343 EYC38099.1 JARK01001459 EYB99276.1 EYC03125.1 JARK01001421 EYC04912.1 EYB99275.1 JARK01001346 EYC26445.1 JARK01000109 EYC42952.1
Proteomes
Pfam
Interpro
IPR005135
Endo/exonuclease/phosphatase
+ More
IPR036691 Endo/exonu/phosph_ase_sf
IPR000477 RT_dom
IPR029052 Metallo-depent_PP-like
IPR000719 Prot_kinase_dom
IPR017441 Protein_kinase_ATP_BS
IPR008271 Ser/Thr_kinase_AS
IPR011009 Kinase-like_dom_sf
IPR002213 UDP_glucos_trans
IPR001507 ZP_dom
IPR018422 Cation/H_exchanger_CPA1
IPR006153 Cation/H_exchanger
IPR004709 NaH_exchanger
IPR019430 7TM_GPCR_serpentine_rcpt_Srx
IPR017452 GPCR_Rhodpsn_7TM
IPR036691 Endo/exonu/phosph_ase_sf
IPR000477 RT_dom
IPR029052 Metallo-depent_PP-like
IPR000719 Prot_kinase_dom
IPR017441 Protein_kinase_ATP_BS
IPR008271 Ser/Thr_kinase_AS
IPR011009 Kinase-like_dom_sf
IPR002213 UDP_glucos_trans
IPR001507 ZP_dom
IPR018422 Cation/H_exchanger_CPA1
IPR006153 Cation/H_exchanger
IPR004709 NaH_exchanger
IPR019430 7TM_GPCR_serpentine_rcpt_Srx
IPR017452 GPCR_Rhodpsn_7TM
Gene 3D
ProteinModelPortal
D7F179
D7F176
A0A2H1WF27
D7F177
D7F178
A0A023EXV2
+ More
A0A2W1BZV1 A0A2H1VS24 A0A069DZM5 D7F157 D7F164 D7F165 D7F166 D7F160 A0A016S1D2 A0A016TK11 A0A016SG46 A0A016W4R1 E3NC15 E3MH09 A0A016WPY4 A0A016WCB8 A0A016ULU9 A0A016VEC1 A0A016VCD9 A0A016T590 A0A016T7J7 A0A016VCU3 A0A016VBP7 A0A0K0FAW8 A0A016V6A2 E3NGJ2 A0A016S5B2 E3N0F4 A0A016VVN3 A0A016S795 A0A016WA79 A0A016UN79 Q9TZX4 A0A016SX67 K7IG35 A0A2H2J8I4 A0A016V419 G0MZ46 A0A2H2JFN0 K7ICT2 K7ICT1 A0A016RUR3 A0A016RV73 K7H0R0 K7IBZ6 K7H0Q9 K7I2A3 K7IBZ5 A0A016SLV8 K7IG36 K7HDM2 K7HDM3 A0A016RRX8 E3NJQ7 A0A016THC6 A0A016U352 A0A016U3L3 A0A016T4C4 A0A016SGC3 A0A016SVA2 K7I2A2 A0A016SW37 A0A016W1X2 A0A016WYN9 A0A016SPR9 A0A016SCZ8 K7H0Q8 A0A016SCL7 A0A016VLP0 A0A016SY13 A0A016U6B7 A0A016VCK8 A0A016U123 K7I2A1 A0A016X207 A0A016WFS9 A0A016SCT9 A0A016VT25 A0A2W5U8K4 A0A016UJS4 A0A016WE57 A0A016T9D3 A0A016TK92 A0A016TQJ1 A0A016T9A5 A0A016VGA0 A0A016WUN2
A0A2W1BZV1 A0A2H1VS24 A0A069DZM5 D7F157 D7F164 D7F165 D7F166 D7F160 A0A016S1D2 A0A016TK11 A0A016SG46 A0A016W4R1 E3NC15 E3MH09 A0A016WPY4 A0A016WCB8 A0A016ULU9 A0A016VEC1 A0A016VCD9 A0A016T590 A0A016T7J7 A0A016VCU3 A0A016VBP7 A0A0K0FAW8 A0A016V6A2 E3NGJ2 A0A016S5B2 E3N0F4 A0A016VVN3 A0A016S795 A0A016WA79 A0A016UN79 Q9TZX4 A0A016SX67 K7IG35 A0A2H2J8I4 A0A016V419 G0MZ46 A0A2H2JFN0 K7ICT2 K7ICT1 A0A016RUR3 A0A016RV73 K7H0R0 K7IBZ6 K7H0Q9 K7I2A3 K7IBZ5 A0A016SLV8 K7IG36 K7HDM2 K7HDM3 A0A016RRX8 E3NJQ7 A0A016THC6 A0A016U352 A0A016U3L3 A0A016T4C4 A0A016SGC3 A0A016SVA2 K7I2A2 A0A016SW37 A0A016W1X2 A0A016WYN9 A0A016SPR9 A0A016SCZ8 K7H0Q8 A0A016SCL7 A0A016VLP0 A0A016SY13 A0A016U6B7 A0A016VCK8 A0A016U123 K7I2A1 A0A016X207 A0A016WFS9 A0A016SCT9 A0A016VT25 A0A2W5U8K4 A0A016UJS4 A0A016WE57 A0A016T9D3 A0A016TK92 A0A016TQJ1 A0A016T9A5 A0A016VGA0 A0A016WUN2
Ontologies
PANTHER
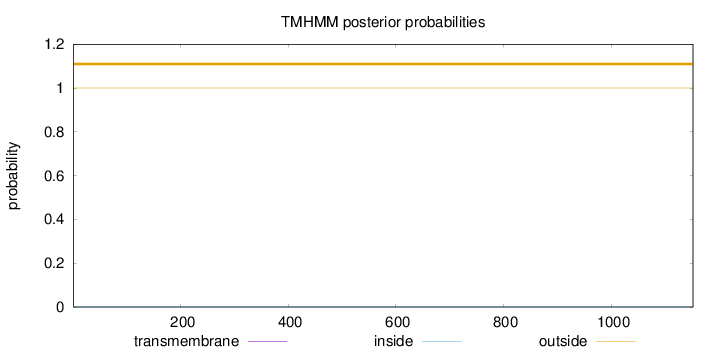
Topology
Length:
1152
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0005
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00000
outside
1 - 1152
Population Genetic Test Statistics
Pi
10.348894
Theta
30.16349
Tajima's D
0.653805
CLR
34.250886
CSRT
0.56157192140393
Interpretation
Uncertain
