Pre Gene Modal
BGIBMGA012165
Annotation
PREDICTED:_WD_repeat-containing_protein_74_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.439 Nuclear Reliability : 2.089
Sequence
CDS
ATGGTGGTTAATGAAGATTCTCAGTTAGACATATTCGTGGGTAGTACGATTGGATCGTTCAAGCATGTAAAATATTACACGGACAACACGAAAAACAACAAGAAATGCATAGAAAACTTGGTCGACGTTAAATCTTTACAGAAAGGCGAAGGTATAACTTGCATGGAATGGGGTAACAAAGAACAAACTGAAATTTTGATTGGAAAGAAAAACCAACAGATACAAGTTTACCACACTTTACAAGGAAACAAGTGTATTTATACAGCCGACTATGCAGAAGGACATGTCGTAGGTCTGGGGAAAGTGAACAATACATTGGTATCAGCCGTGTCAAGTGGCACCGTCAGAGTATGGGGCGATGATGCGAATGAAGATATTTTCACTGGCGGGAAGCTGGATAGGATGAGGGTCTGCAGCGATGATACATTATTTGCTACAGGCGGCGAAGAAAACGATTTGAAAGTTTGGCGCGTCGGCGAGTCCCAGCCAACGTTCTCTGCCAAGAACTTGGCCCACGATTGGCTCCAACTACGCCAGCCGGTCTGGGTGACCGACCTGGTGTTTGTCCCCGGCGAGGGGGGCAGGGTGGTAGCCGTCTGCTCCCGACACGGCTACGTGAGATTGTACGACACTCGCTCGCAAAGAAGGCCGGTCCAGAACGTGACCTTCGATCTGGCGGCCACTTGCATCGCGCCCAGCATCAGCACGCGTCTGGCGTTGGTGGGGTTCGCCCGGGGACAGCTCCGGCAGGTGGAACTGCGGCTGGGGCGCCCCGACAAGGGGTACAAGGGGGGTGCGGGGGCGGTGACGGGCGCGGGGGCGGGGGGCGGCCGCGTGCTCAGTACCTCCCTGGACAGACATCTATACGTGCACCGATACAGCGACAAGGAACTCGTTCATAAGGGACCCCGGACCGGTCAGCAAGAGACCCAAACCCTCGCTGGAGGGCAGCACCGAAGTGGTGGAAGACACGGAAGCGGCCATAGCGAAGCTCCTCCGCAGCACGGAGAAGACCAGGAAGAGGAGGGAGCAGCGGAAGCGGGAGAAGAAAAAGAAGAGCGTCTTTCATAA
Protein
MVVNEDSQLDIFVGSTIGSFKHVKYYTDNTKNNKKCIENLVDVKSLQKGEGITCMEWGNKEQTEILIGKKNQQIQVYHTLQGNKCIYTADYAEGHVVGLGKVNNTLVSAVSSGTVRVWGDDANEDIFTGGKLDRMRVCSDDTLFATGGEENDLKVWRVGESQPTFSAKNLAHDWLQLRQPVWVTDLVFVPGEGGRVVAVCSRHGYVRLYDTRSQRRPVQNVTFDLAATCIAPSISTRLALVGFARGQLRQVELRLGRPDKGYKGGAGAVTGAGAGGGRVLSTSLDRHLYVHRYSDKELVHKGPRTGQQETQTLAGGQHRSGGRHGSGHSEAPPQHGEDQEEEGAAEAGEEKEERLS
Summary
Uniprot
A0A2A4JIX2
H9JRK5
A0A2H1VH07
S4PBE3
A0A0N1INA5
A0A212ESE2
+ More
A0A0N0PED3 A0A1B6CVN9 A0A1S3DJJ9 A0A1S4EP51 A0A0V0GB34 A0A023FA33 A0A224XT71 A0A0P4VGA4 R4G809 A0A1B6LEY6 E0VVJ3 A0A026WWE3 R7TR62 A0A1B6F062 K7ITM1 A0A232EUT7 Q6DDA2 A2VD86 A0A1L8GJW2 A0A0P6GYT7 A0A0P5HI51 A0A0P6F079 A0A0P6C0L7 A0A0P5PYK3 A0A0P6I7W4 A0A0P5YK56 A0A067R7H9 A0A0P5B1K3 E9FX62 A0A0M8ZUE6 A0A1B0CZG1 A0A0P5L2T1 A0A0P5YCW6 A0A2J7Q4B7 A0A0P6DYM2 E2AIU1 V9KY75 A0A1S3HAN6 U5EW31 K7FDE6 C3XZK4 A0A151MTE9 A0A1U7SMK4 T1ITQ1 A0A210PSD0 A0A088A269 E2BA52 A0A310SGS2 V9L1Q8 A0A0A9WUP3 A0A0K8S7G9 A0A2D0SP32 A0A2D0SPP0 W5UDN5 H3A1N8 K1P1Q8 A0A2J7Q4C1 A0A0L7RFR9 A0A146LAP7 M7B1B6 A0A2L2Y155 K1QJ98 A0A0P5EA88 A0A0P5YMU0 T1E434 A0A2A3E9Q5 T1J0T6 A0A0P5RQA6
A0A0N0PED3 A0A1B6CVN9 A0A1S3DJJ9 A0A1S4EP51 A0A0V0GB34 A0A023FA33 A0A224XT71 A0A0P4VGA4 R4G809 A0A1B6LEY6 E0VVJ3 A0A026WWE3 R7TR62 A0A1B6F062 K7ITM1 A0A232EUT7 Q6DDA2 A2VD86 A0A1L8GJW2 A0A0P6GYT7 A0A0P5HI51 A0A0P6F079 A0A0P6C0L7 A0A0P5PYK3 A0A0P6I7W4 A0A0P5YK56 A0A067R7H9 A0A0P5B1K3 E9FX62 A0A0M8ZUE6 A0A1B0CZG1 A0A0P5L2T1 A0A0P5YCW6 A0A2J7Q4B7 A0A0P6DYM2 E2AIU1 V9KY75 A0A1S3HAN6 U5EW31 K7FDE6 C3XZK4 A0A151MTE9 A0A1U7SMK4 T1ITQ1 A0A210PSD0 A0A088A269 E2BA52 A0A310SGS2 V9L1Q8 A0A0A9WUP3 A0A0K8S7G9 A0A2D0SP32 A0A2D0SPP0 W5UDN5 H3A1N8 K1P1Q8 A0A2J7Q4C1 A0A0L7RFR9 A0A146LAP7 M7B1B6 A0A2L2Y155 K1QJ98 A0A0P5EA88 A0A0P5YMU0 T1E434 A0A2A3E9Q5 T1J0T6 A0A0P5RQA6
Pubmed
EMBL
NWSH01001406
PCG71352.1
BABH01031773
BABH01031774
ODYU01002508
SOQ40097.1
+ More
GAIX01004661 JAA87899.1 KQ458863 KPJ04711.1 AGBW02012817 OWR44400.1 KQ459988 KPJ18780.1 GEDC01019844 JAS17454.1 GECL01000920 JAP05204.1 GBBI01000929 JAC17783.1 GFTR01005265 JAW11161.1 GDKW01003373 JAI53222.1 ACPB03004692 GAHY01001792 JAA75718.1 GEBQ01017719 JAT22258.1 DS235812 EEB17399.1 KK107105 EZA59449.1 AMQN01012467 AMQN01012468 KB309539 ELT93981.1 GECZ01026181 JAS43588.1 NNAY01002087 OXU22120.1 BC077693 CR761697 AAH77693.1 CAJ83923.1 BC129603 AAI29604.1 CM004472 OCT84103.1 GDIQ01047929 JAN46808.1 GDIQ01227162 JAK24563.1 GDIQ01067615 JAN27122.1 GDIP01006625 JAM97090.1 GDIQ01121646 JAL30080.1 GDIQ01015309 JAN79428.1 GDIP01070656 JAM33059.1 KK852655 KDR19313.1 GDIP01195396 JAJ28006.1 GL732526 EFX88020.1 KQ435881 KOX69813.1 AJVK01020757 GDIQ01200812 GDIQ01193265 JAK50913.1 GDIP01059514 JAM44201.1 NEVH01018385 PNF23432.1 GDIQ01070766 JAN23971.1 GL439905 EFN66620.1 JW871124 AFP03642.1 GANO01001605 JAB58266.1 AGCU01052713 GG666475 EEN66525.1 AKHW03005097 KYO27786.1 JH431494 NEDP02005528 OWF39391.1 GL446664 EFN87461.1 KQ764001 OAD54650.1 JW872334 AFP04852.1 GBHO01032488 JAG11116.1 GBRD01016599 JAG49227.1 JT412776 AHH39990.1 AFYH01015686 AFYH01015687 JH821810 EKC17777.1 PNF23433.1 KQ414601 KOC69680.1 GDHC01013225 JAQ05404.1 KB586437 EMP25858.1 IAAA01006602 LAA01919.1 JH818661 EKC28945.1 GDIP01144346 JAJ79056.1 GDIP01056952 JAM46763.1 GAAZ01002941 JAA95002.1 KZ288311 PBC28450.1 AFFK01020674 GDIQ01098450 JAL53276.1
GAIX01004661 JAA87899.1 KQ458863 KPJ04711.1 AGBW02012817 OWR44400.1 KQ459988 KPJ18780.1 GEDC01019844 JAS17454.1 GECL01000920 JAP05204.1 GBBI01000929 JAC17783.1 GFTR01005265 JAW11161.1 GDKW01003373 JAI53222.1 ACPB03004692 GAHY01001792 JAA75718.1 GEBQ01017719 JAT22258.1 DS235812 EEB17399.1 KK107105 EZA59449.1 AMQN01012467 AMQN01012468 KB309539 ELT93981.1 GECZ01026181 JAS43588.1 NNAY01002087 OXU22120.1 BC077693 CR761697 AAH77693.1 CAJ83923.1 BC129603 AAI29604.1 CM004472 OCT84103.1 GDIQ01047929 JAN46808.1 GDIQ01227162 JAK24563.1 GDIQ01067615 JAN27122.1 GDIP01006625 JAM97090.1 GDIQ01121646 JAL30080.1 GDIQ01015309 JAN79428.1 GDIP01070656 JAM33059.1 KK852655 KDR19313.1 GDIP01195396 JAJ28006.1 GL732526 EFX88020.1 KQ435881 KOX69813.1 AJVK01020757 GDIQ01200812 GDIQ01193265 JAK50913.1 GDIP01059514 JAM44201.1 NEVH01018385 PNF23432.1 GDIQ01070766 JAN23971.1 GL439905 EFN66620.1 JW871124 AFP03642.1 GANO01001605 JAB58266.1 AGCU01052713 GG666475 EEN66525.1 AKHW03005097 KYO27786.1 JH431494 NEDP02005528 OWF39391.1 GL446664 EFN87461.1 KQ764001 OAD54650.1 JW872334 AFP04852.1 GBHO01032488 JAG11116.1 GBRD01016599 JAG49227.1 JT412776 AHH39990.1 AFYH01015686 AFYH01015687 JH821810 EKC17777.1 PNF23433.1 KQ414601 KOC69680.1 GDHC01013225 JAQ05404.1 KB586437 EMP25858.1 IAAA01006602 LAA01919.1 JH818661 EKC28945.1 GDIP01144346 JAJ79056.1 GDIP01056952 JAM46763.1 GAAZ01002941 JAA95002.1 KZ288311 PBC28450.1 AFFK01020674 GDIQ01098450 JAL53276.1
Proteomes
UP000218220
UP000005204
UP000053268
UP000007151
UP000053240
UP000079169
+ More
UP000015103 UP000009046 UP000053097 UP000014760 UP000002358 UP000215335 UP000186698 UP000027135 UP000000305 UP000053105 UP000092462 UP000235965 UP000000311 UP000085678 UP000007267 UP000001554 UP000050525 UP000189705 UP000242188 UP000005203 UP000008237 UP000221080 UP000008672 UP000005408 UP000053825 UP000031443 UP000242457
UP000015103 UP000009046 UP000053097 UP000014760 UP000002358 UP000215335 UP000186698 UP000027135 UP000000305 UP000053105 UP000092462 UP000235965 UP000000311 UP000085678 UP000007267 UP000001554 UP000050525 UP000189705 UP000242188 UP000005203 UP000008237 UP000221080 UP000008672 UP000005408 UP000053825 UP000031443 UP000242457
PRIDE
Interpro
IPR001680
WD40_repeat
+ More
IPR037379 WDR74/Nsa1
IPR036322 WD40_repeat_dom_sf
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR017986 WD40_repeat_dom
IPR011171 GMF
IPR029006 ADF-H/Gelsolin-like_dom_sf
IPR002108 ADF-H
IPR001320 Iontro_rcpt
IPR001254 Trypsin_dom
IPR009003 Peptidase_S1_PA
IPR033116 TRYPSIN_SER
IPR037379 WDR74/Nsa1
IPR036322 WD40_repeat_dom_sf
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR017986 WD40_repeat_dom
IPR011171 GMF
IPR029006 ADF-H/Gelsolin-like_dom_sf
IPR002108 ADF-H
IPR001320 Iontro_rcpt
IPR001254 Trypsin_dom
IPR009003 Peptidase_S1_PA
IPR033116 TRYPSIN_SER
Gene 3D
ProteinModelPortal
A0A2A4JIX2
H9JRK5
A0A2H1VH07
S4PBE3
A0A0N1INA5
A0A212ESE2
+ More
A0A0N0PED3 A0A1B6CVN9 A0A1S3DJJ9 A0A1S4EP51 A0A0V0GB34 A0A023FA33 A0A224XT71 A0A0P4VGA4 R4G809 A0A1B6LEY6 E0VVJ3 A0A026WWE3 R7TR62 A0A1B6F062 K7ITM1 A0A232EUT7 Q6DDA2 A2VD86 A0A1L8GJW2 A0A0P6GYT7 A0A0P5HI51 A0A0P6F079 A0A0P6C0L7 A0A0P5PYK3 A0A0P6I7W4 A0A0P5YK56 A0A067R7H9 A0A0P5B1K3 E9FX62 A0A0M8ZUE6 A0A1B0CZG1 A0A0P5L2T1 A0A0P5YCW6 A0A2J7Q4B7 A0A0P6DYM2 E2AIU1 V9KY75 A0A1S3HAN6 U5EW31 K7FDE6 C3XZK4 A0A151MTE9 A0A1U7SMK4 T1ITQ1 A0A210PSD0 A0A088A269 E2BA52 A0A310SGS2 V9L1Q8 A0A0A9WUP3 A0A0K8S7G9 A0A2D0SP32 A0A2D0SPP0 W5UDN5 H3A1N8 K1P1Q8 A0A2J7Q4C1 A0A0L7RFR9 A0A146LAP7 M7B1B6 A0A2L2Y155 K1QJ98 A0A0P5EA88 A0A0P5YMU0 T1E434 A0A2A3E9Q5 T1J0T6 A0A0P5RQA6
A0A0N0PED3 A0A1B6CVN9 A0A1S3DJJ9 A0A1S4EP51 A0A0V0GB34 A0A023FA33 A0A224XT71 A0A0P4VGA4 R4G809 A0A1B6LEY6 E0VVJ3 A0A026WWE3 R7TR62 A0A1B6F062 K7ITM1 A0A232EUT7 Q6DDA2 A2VD86 A0A1L8GJW2 A0A0P6GYT7 A0A0P5HI51 A0A0P6F079 A0A0P6C0L7 A0A0P5PYK3 A0A0P6I7W4 A0A0P5YK56 A0A067R7H9 A0A0P5B1K3 E9FX62 A0A0M8ZUE6 A0A1B0CZG1 A0A0P5L2T1 A0A0P5YCW6 A0A2J7Q4B7 A0A0P6DYM2 E2AIU1 V9KY75 A0A1S3HAN6 U5EW31 K7FDE6 C3XZK4 A0A151MTE9 A0A1U7SMK4 T1ITQ1 A0A210PSD0 A0A088A269 E2BA52 A0A310SGS2 V9L1Q8 A0A0A9WUP3 A0A0K8S7G9 A0A2D0SP32 A0A2D0SPP0 W5UDN5 H3A1N8 K1P1Q8 A0A2J7Q4C1 A0A0L7RFR9 A0A146LAP7 M7B1B6 A0A2L2Y155 K1QJ98 A0A0P5EA88 A0A0P5YMU0 T1E434 A0A2A3E9Q5 T1J0T6 A0A0P5RQA6
PDB
5SUI
E-value=1.0756e-05,
Score=117
Ontologies
GO
PANTHER
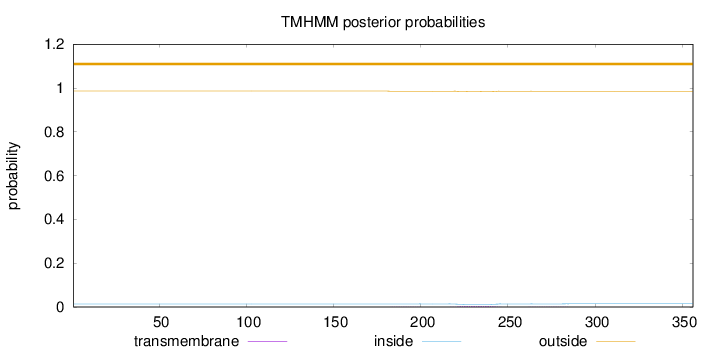
Topology
Length:
356
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.10822
Exp number, first 60 AAs:
0.00178
Total prob of N-in:
0.01393
outside
1 - 356
Population Genetic Test Statistics
Pi
22.150734
Theta
202.349113
Tajima's D
1.188931
CLR
0.199624
CSRT
0.712314384280786
Interpretation
Uncertain
